Strain
Human_OC43_AY391777
(Region: United Kingdom; Strain: Human coronavirus OC43, complete genome.; Date: 1967)
Gene
HE protein
Description
Annotated in NCBI,
HE protein
Location
GenBank Accession
Full name
Hemagglutinin-esterase
Alternative Name
E3 glycoprotein
Sequence
CDS
ATGTTTTTGCTTCCTAGATTTATTCTAGTTAGCTGCATAATTGGTAGCTTAGGTTTTTACAACCCTCCTACCAATGTTGTTTCGCATGTAAATGGAGATTGGTTTTTATTTGGTGACAGTCGTTCAGATTGTAATCATATTGTTAATATCAACCCCCATAATTATTCTTATATGGACCTTAATCCTGTTCTGTGTGATTCTGGTAAAATATCATCTAAAGCTGGCAACTCCATTTTTAGGAGTTTTCACTTTACCGATTTTTATAATTACACAGGCGAAGGTCAACAAATTATTTTTTATGAGGGTGTTAATTTTACGCCTTATCATGCCTTTAAATGCAACCGTTCTGGTAGTAATGATATTTGGATGCAGAATAAAGGCTTGTTTTATACTCAGGTTTATAAGAATATGGCTGTGTATCGCAGCCTTACTTTTGTTAATGTACCATATGTTTATAATGGCTCCGCACAAGCTACAGCTCTTTGTAAATCTGGTAGTTTAGTCCTTAATAACCCTGCATATATAGCTCCTCAAGCTAACTCTGGGGATTATTATTATAAGGTTGAAGCTGATTTTTATTTGTCAGGTTGTGACGAGTATATCGTACCACTTTGTATTTTTAACGGCAAGTTTTTGTCGAATACAAAGTATTATGATGATAGTCAATATTATTTTAATAAAGACACTGGTGTTATTTATGGTCTCAATTCTACAGAAACCATTACCACTGGTTTTGATCTTAATTGTTATTATTTAGTTTTACCCTCTGGTAATTATTTAGCCATTTCAAATGAGCTATTGTTAACTGTTCCTACGAAAGCAATCTGTCTTAATAAGCGTAAGGATTTTACGCCTGTACAGGTTGTTGATTCGCGGTGGAACAATGCCAGGCAGTCTGATAACATGACGGCGGTTGCTTGTCAACCTCCGTACTGTTATTTTCGTAATTCTACTACCAACTATGTTGGTGTTTATGATATTAATCATGGAGATGCTGGTTTTACTAGCATACTTAGTGGTTTGTTATATAATTCACCTTGTTTTTCGCAGCAAGGCGTTTTTAGGTATGATAATGTTAGCAGTGTCTGGCCTCTCTACCCCTATGGCAGATGTCCCACTGCTGCTGATATTAATATCCCTGATTTACCCATTTGTGTGTATGATCCGCTACCAGTTATTTTGCTTGGCATTCTTTTGGGCGTTGCGATTGTAATTATTGTAGTTTTGTTGTTATATTTTATGGTGGATAATGTTACTAGGCTGCATGATGCTTAG
Protein
MFLLPRFILVSCIIGSLGFYNPPTNVVSHVNGDWFLFGDSRSDCNHIVNINPHNYSYMDLNPVLCDSGKISSKAGNSIFRSFHFTDFYNYTGEGQQIIFYEGVNFTPYHAFKCNRSGSNDIWMQNKGLFYTQVYKNMAVYRSLTFVNVPYVYNGSAQATALCKSGSLVLNNPAYIAPQANSGDYYYKVEADFYLSGCDEYIVPLCIFNGKFLSNTKYYDDSQYYFNKDTGVIYGLNSTETITTGFDLNCYYLVLPSGNYLAISNELLLTVPTKAICLNKRKDFTPVQVVDSRWNNARQSDNMTAVACQPPYCYFRNSTTNYVGVYDINHGDAGFTSILSGLLYNSPCFSQQGVFRYDNVSSVWPLYPYGRCPTAADINIPDLPICVYDPLPVILLGILLGVAIVIIVVLLLYFMVDNVTRLHDA
Summary
Function
Structural protein that makes short spikes at the surface of the virus. Contains receptor binding and receptor-destroying activities. Mediates de-O-acetylation of N-acetyl-4-O-acetylneuraminic acid, which is probably the receptor determinant recognized by the virus on the surface of erythrocytes and susceptible cells. This receptor-destroying activity is important for virus release as it probably helps preventing self-aggregation and ensures the efficient spread of the progeny virus from cell to cell. May serve as a secondary viral attachment protein for initiating infection, the spike protein being the major one. May become a target for both the humoral and the cellular branches of the immune system.
Catalytic Activity
H2O + N-acetyl-4-O-acetylneuraminate = acetate + H(+) + N-acetylneuraminate
H2O + N-acetyl-9-O-acetylneuraminate = acetate + H(+) + N-acetylneuraminate
H2O + N-acetyl-9-O-acetylneuraminate = acetate + H(+) + N-acetylneuraminate
Subunit
Homodimer; disulfide-linked. Forms a complex with the M protein in the pre-Golgi. Associates then with S-M complex to form a ternary complex S-M-HE.
Similarity
Belongs to the influenza type C/coronaviruses hemagglutinin-esterase family.
Feature
chain Hemagglutinin-esterase
Uniprot
EC Number
3.1.1.53
Pubmed
EMBL
Proteomes
Interpro
SUPFAM
SSF49818
SSF49818
ProteinModelPortal
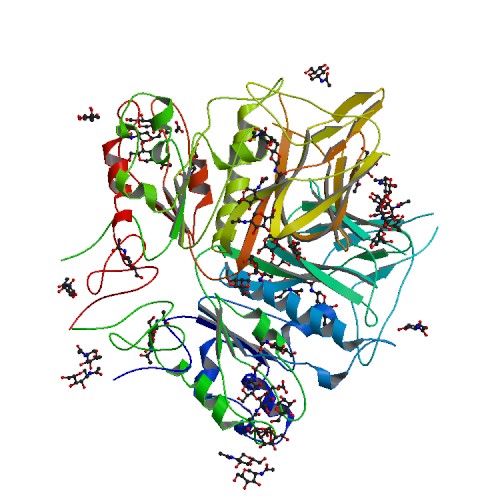
PDB
5N11
E-value=0.0 Score= 729 Identity=96.49%
Cov(Q)=87.26% Cov(P)=98.40%
3D Structure
Ontologies
GO
Subcellular Location
From MSLVP
Single-Pass Membrane
From Uniprot
Virion membrane
In infected cells becomes incorporated into the envelope of virions during virus assembly at the endoplasmic reticulum and cis Golgi. However, some may escape incorporation into virions and subsequently migrate to the cell surface. With evidence from 1 publications.
Host cell membrane In infected cells becomes incorporated into the envelope of virions during virus assembly at the endoplasmic reticulum and cis Golgi. However, some may escape incorporation into virions and subsequently migrate to the cell surface. With evidence from 1 publications.
Host cell membrane In infected cells becomes incorporated into the envelope of virions during virus assembly at the endoplasmic reticulum and cis Golgi. However, some may escape incorporation into virions and subsequently migrate to the cell surface. With evidence from 1 publications.
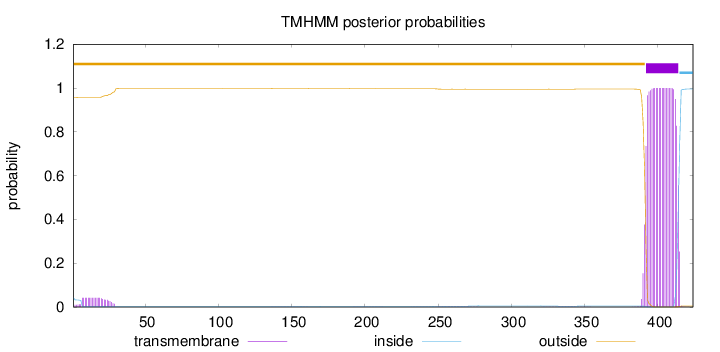
Topology
Length:
424
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.83313
Exp number, first 60 AAs:
0.88103
Total prob of N-in:
0.04249
outside
1 - 391
TMhelix
392 - 414
inside
415 - 424