Strain
Bat_KU762338
(Region: China; Strain: Rousettus bat coronavirus GCCDC1; Date: 2014/5/28)
Gene
spike protein
Description
Annotated in NCBI,
spike protein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGTTTCGGACGTTGCTCTTAGCGGCGTTGTTTAACGCCTGCTGGTGTGCGCGTCCGACTTGTATTAAGCCTACTGAAACATGGGGCACGCCTTCATTTACGGGTGTGGAGTATGTACCACATAACACTACCTATGTCTCAGTACCGCTTAACAAGCTATCATGCTTATTACCAGACCCTTATATGGCACACTCAGGACAGACTGTGAGACAAAAGTTATACATGGGCAATACCTCCAATACGCTTGTATACCCAGTCACACCACCAATGTTCAACCTTACATATGGCAATGTGACACCTGGTGTGTACAACACGTCTTTTCTACCAGTCTTTGACGGGCTCCTTGTGCATACCTACATGAATCGGTTTGCACACTTGGATAACCCTAATAGGACATGTCAAGAGCCATTTGGCGTTGTGTTTGGCACTACCTTTGAGCAAGACCGTATTGCTATGGTCATAATTGCTCCAGGTGAGTTTGGTATGTGGGGTCAGGTTAACCGGCCTAACACTACTTTTGTTCATGTAGTGGCATGTAGTAATGCTACCATTTGTGCCTACCCTATGTTCAATAGGTGGGGACCAGCTGGTAGCATACATGCCGAAAACTCCTTTGTTGAGCACAACTCCTCGTGCTTTTTGAATGACACTTTTGAGATTCCAATGGGTACATCTCGTGTCAATTTAGCTTTTAGGTTTCAAGACGGCAACCTTTTACTGTACCATACAGCTTGGCTGCCGACTGAAAACTATACTTTAAGTGGTGACTACCCCCTACGTTACGCACGTAGTGTTGGTGTAGGCTCCAATTTACCGTTTGCTCAATTCTTTCAGTCTGTCGCTCGTGCACGTGATAGTGCATGCGCTTTCCTACATAATAATTTGTACATAGCCCCTGTACAGCCTAAAGAGCTGCTTGTTAAGTACAATGACCAAGGCATGCCTGTAGAAATTGCAGACTGCTCTGCTGATTCTACACAAGAGCTATATTGTATTACTGGCACCTTCACACCAAGTGTCGGTGTTTATCAATTAAGTCGCTATCGCGCTGAAGCCAAAAAGCTTGTGCAAGTTACACAGCAAGAAGACTCCTGCGCGATACCGTATACTACTATTCTAGAACCACCCTCACCGGCTGCGTGGGTGCGTGCTACCATTTCTAATTGTACTTTCGACTTTGAATCGCTACTGCGGACATTGCCGACATACAATTTGAAGTGCTATGGCATCTCACCTGCTCGACTTAGTACTATGTGCTATGCTGGGGTGACGCTAGATATTTTTAAACTAAACACCACCCACTTGTCCAACATGCTTGGATCTGTGCCGGATGCTGTTAGTATTTACAATTATGCCTTGCCTAGCAACTTTTATGGTTGCGTGCATGCTTATCATCTTAATAGTACTACTCCATATGCAGTGGCTGTACCACCTGGCGCTTACCCAATTAAACCAGGTGGCAGACAGTTGTTTAACAGCTTTGTGTCTCAAGTCTTAGACTCACCAACGTCACAGTGCACCCCTGCTAATTGCATGGGTGTTGTTGTTATAGGGTTAACACCAGCTTCTGGTTCTAACCTCGTGTGTCCAAAGGCTAACGATACACAAGTTATCGAAGGTCAGTGTGTGAAGTACAACTTTTATGGCTATGCAGGCACCGGTGTTATAAACCAGAGTGACCTTGCCATACCCAACAACAAGTTGTTTGTTACTAGCAAATCAGGAGCTGTCCTTGCCGTACGTGCAGGTGACAAAGTTTACAGCATCACACCTTGTGTGTCAGTGCCCATTTCAGTTGGTTATGACCCTGGACATCAGCGTGCGCTTGTGTTCAACGGTCTCGACTGTAGTGCGCGCGCGAATGCTGTAAGCATGCCAGCTTCCGAATATTGGACCGCAGCCGCTAGCACTACTGCTCGCGGTTCTGAGCCTGTATTAGACACACCGTCTGGTTGCGTCTACAATGTTAATAACTGCACTACACACACAGTTAGTGTTTGTGAAATGCCTATTGGTAACAGCCTTTGCCTCGTCAGCAACTTTACGTGTTCTGACGTGGCAACAGCTAGCCTTAGTCCAAATTTGTTAAGCTTGGTTGTATACGACCCCACTGATGCGGGACTTAAAGTTCTAACACCGGTGTACTGGGTCAGTATACCCACCAACTTTACACTAGCTGCAACAACTGAGTACATTCAAACCACTGCACCCAAAGTCAATGTTGACTGTGTTAAGTACCTATGTGGCGACTCTGAGCGCTGCATAGATGTGCTGTCACAATATGGGGCTTTCTGTGAAGACGTCAACAAGGCTCTTGCCGACGTCAGTGCTATTATTGATTCATCTATGGTGACTATGGTTTCAGAGCTTACTGCCGGTGTGATGTGGAGTGAAACACCTCAAGCTAATGTCGGCTCATATAACTTTTCCGGTCTTATGGGCTGTCTAGGTTCCAACTGTCAGGAGAAGCAGTATCGTTCTGCTATCTCAGACTTGTTGTATAATAAAGTTAAGGTAGCAGACCCAGGGTTTATGGGTGCTTACCAGAAATGTATTGATGAGCAGTGGGGTGGCAGTGTGAGAGACCTTATTTGCACTCAGACATTTAATGGCATCTCAGTGTTACCGCCCATTGTCTCTCCTGGCATGCAAGCCTTGTATACATCTTTATTAGTAGGTGCTGTTGCTTCATCTGGCTACACCTTTGGCATTACGTCCGTTGGTGTCATACCATTTGCCACACAGCTCCAATTTAGACTTAATGGCCTTGGTGTTACCACCAATGTGCTCATGGAAAACCAAAAACTCATTGCCAACGCATTCAACAATGCCCTCACAGGCATACAGAAGGGTTTTGACGCTACCAACATGGCTTTAGCCAAAATGCAGAGTGTCATCAACCAGCATGCGCAGCAATTGAGTACTTTAGTTGACCAATTGGGCAACTCTTTTGGTGCTATCTCCTCATCCATCAATGAAATCTTTAGTCGGCTAGATGAATTAGAAGCCAATGCACAAGTAGATCGCCTTATCAATGGGCGTATGGTTGTCTTGAACACCTACGTCACTCAGCTGCTCATTAGAGCTTCTGAGGTTAAGGCACAAGCTGCGTTGGCTAGTCAAAAGATAAGTGAGTGTGTCAAAGCACAATCCCAGCGTAATGACTTTTGCGGTAATGGCACACATGTGTTGTCCATACCTCAAATGGCACCAAATGGTGTGTTGTTCTTGCACTACTCCTATCAACCTACAGCTTACAATTTAGTGCGTACTGCCGCTGGCCTTTGTTTTAATGACACTGGCTATGCACCTCTAGGAGGTTTGTTTGTGTTGCCTAACAACACAGACAGGTGGCTATTCACTAAAATGAGCTTTTACGATCCCGTAAACATCTCAGTTTCTAATACGCAGGTCCTTGCGGCTTGTGGTCTTAATTATTCTAGTGTCAACTACACCGTCCTTGAGCCGGCGGTTGACACTAGCTCCTTTAATTTTACAGAAGAGTTTGAAAAGTGGTACGTCAACCAGAGTCATATTTTTAACAACACCTTTAACGCCAGCGCCTTTAACTTTTCCCTTGTGGACGTTAATGAGCAGCTTGCGATTCTTACAGATGTTGTCAACCAGTTAAACCAGTCTTACATAGACCTCAAGCAGTTGGGCACCTATGAGTACACGGTCAAATGGCCGTGGTACGTATGGCTGGGTATGATTGCAGGACTAGTAGGCCTTGTGCTAGCGGTAGTTCTGCTGCTTTGTATGACTAATTGCTGCTCGTGTGCTCGCGGTGTTTGCTCTTGCAAGTCGTGTGCTTATGAGGAGCATGAGGATGTGTATCCTGCTGTTCGTGTGCATGGAAAACGAACGGCATGA
Protein
MFRTLLLAALFNACWCARPTCIKPTETWGTPSFTGVEYVPHNTTYVSVPLNKLSCLLPDPYMAHSGQTVRQKLYMGNTSNTLVYPVTPPMFNLTYGNVTPGVYNTSFLPVFDGLLVHTYMNRFAHLDNPNRTCQEPFGVVFGTTFEQDRIAMVIIAPGEFGMWGQVNRPNTTFVHVVACSNATICAYPMFNRWGPAGSIHAENSFVEHNSSCFLNDTFEIPMGTSRVNLAFRFQDGNLLLYHTAWLPTENYTLSGDYPLRYARSVGVGSNLPFAQFFQSVARARDSACAFLHNNLYIAPVQPKELLVKYNDQGMPVEIADCSADSTQELYCITGTFTPSVGVYQLSRYRAEAKKLVQVTQQEDSCAIPYTTILEPPSPAAWVRATISNCTFDFESLLRTLPTYNLKCYGISPARLSTMCYAGVTLDIFKLNTTHLSNMLGSVPDAVSIYNYALPSNFYGCVHAYHLNSTTPYAVAVPPGAYPIKPGGRQLFNSFVSQVLDSPTSQCTPANCMGVVVIGLTPASGSNLVCPKANDTQVIEGQCVKYNFYGYAGTGVINQSDLAIPNNKLFVTSKSGAVLAVRAGDKVYSITPCVSVPISVGYDPGHQRALVFNGLDCSARANAVSMPASEYWTAAASTTARGSEPVLDTPSGCVYNVNNCTTHTVSVCEMPIGNSLCLVSNFTCSDVATASLSPNLLSLVVYDPTDAGLKVLTPVYWVSIPTNFTLAATTEYIQTTAPKVNVDCVKYLCGDSERCIDVLSQYGAFCEDVNKALADVSAIIDSSMVTMVSELTAGVMWSETPQANVGSYNFSGLMGCLGSNCQEKQYRSAISDLLYNKVKVADPGFMGAYQKCIDEQWGGSVRDLICTQTFNGISVLPPIVSPGMQALYTSLLVGAVASSGYTFGITSVGVIPFATQLQFRLNGLGVTTNVLMENQKLIANAFNNALTGIQKGFDATNMALAKMQSVINQHAQQLSTLVDQLGNSFGAISSSINEIFSRLDELEANAQVDRLINGRMVVLNTYVTQLLIRASEVKAQAALASQKISECVKAQSQRNDFCGNGTHVLSIPQMAPNGVLFLHYSYQPTAYNLVRTAAGLCFNDTGYAPLGGLFVLPNNTDRWLFTKMSFYDPVNISVSNTQVLAACGLNYSSVNYTVLEPAVDTSSFNFTEEFEKWYVNQSHIFNNTFNASAFNFSLVDVNEQLAILTDVVNQLNQSYIDLKQLGTYEYTVKWPWYVWLGMIAGLVGLVLAVVLLLCMTNCCSCARGVCSCKSCAYEEHEDVYPAVRVHGKRTA
Summary
Function
Spike protein S1: attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes.
Similarity
Belongs to the betacoronaviruses spike protein family.
Feature
chain Spike protein S1
Uniprot
Proteomes
Interpro
SUPFAM
SSF143587
SSF143587
Gene 3D
ProteinModelPortal
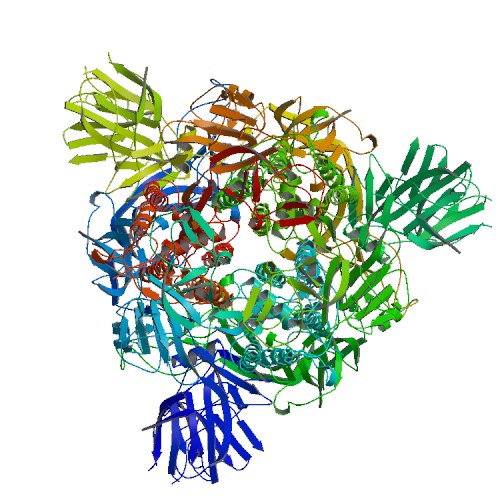
PDB
5XLR
E-value=5e-177 Score= 556 Identity=33.39%
Cov(Q)=88.91% Cov(P)=95.34%
3D Structure
Ontologies
GO
GO:0019031 C:viral envelope
GO:0009405 P:pathogenesis
GO:0016021 C:integral component of membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0020002 C:host cell plasma membrane
GO:0055036 C:virion membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0009405 P:pathogenesis
GO:0016021 C:integral component of membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0020002 C:host cell plasma membrane
GO:0055036 C:virion membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
Subcellular Location
From MSLVP
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Topology
Length:
1290
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
26.09585
Exp number, first 60 AAs:
0.00367
Total prob of N-in:
0.00262
outside
1 - 1229
TMhelix
1230 - 1252
inside
1253 - 1290