Strain
Human_229E_NC_002645
(Region: Germany; Strain: Human coronavirus 229E, complete genome.; Date: 1992)
Gene
surface glycoprotein
Description
Annotated in NCBI,
surface glycoprotein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGTTTGTTTTGCTTGTTGCATATGCCTTGTTGCATATTGCTGGTTGTCAAACTACAAATGGGCTGAACACTAGTTACTCTGTTTGCAACGGCTGTGTTGGTTATTCAGAAAATGTATTTGCTGTTGAGAGTGGTGGTTATATACCCTCCGACTTTGCATTCAATAATTGGTTCCTTCTAACTAATACCTCATCTGTTGTAGATGGTGTTGTGAGGAGTTTTCAGCCTTTGTTGCTTAATTGCTTATGGTCTGTTTCTGGCTTGCGGTTTACTACTGGTTTTGTCTATTTTAATGGTACTGGGAGAGGTGATTGTAAAGGTTTTTCCTCAGATGTTTTGTCTGATGTCATACGTTACAACCTCAATTTTGAAGAAAACCTTAGACGTGGAACCATTTTGTTTAAAACATCTTATGGTGTTGTTGTGTTTTATTGTACCAACAACACTTTAGTTTCAGGTGATGCTCACATACCATTTGGTACAGTTTTGGGCAATTTTTATTGCTTTGTAAATACTACTATTGGCAATGAAACTACGTCTGCTTTTGTGGGTGCACTACCTAAGACAGTTCGTGAGTTTGTTATTTCACGCACAGGACATTTTTATATTAATGGCTATCGCTATTTCACTTTAGGTAATGTAGAAGCCGTTAATTTCAATGTCACTACTGCAGAAACCACTGATTTTTGTACTGTTGCGTTAGCTTCTTATGCTGACGTTTTGGTTAATGTGTCACAAACCTCTATTGCTAATATAATTTATTGCAACTCTGTTATTAACAGACTGAGATGTGACCAGTTGTCCTTTGATGTACCAGATGGTTTTTATTCTACAAGCCCTATTCAATCCGTTGAGCTACCTGTGTCTATTGTGTCGCTACCTGTTTATCATAAACATACGTTTATTGTGTTGTACGTTGACTTCAAACCTCAGAGTGGCGGTGGCAAGTGCTTTAACTGTTATCCTGCTGGTGTTAATATTACACTGGCCAATTTTAATGAAACTAAAGGGCCTTTGTGTGTTGACACATCACACTTCACTACCAAATACGTTGCTGTTTATGCCAATGTTGGTAGGTGGAGTGCTAGTATTAACACGGGAAATTGCCCTTTTTCTTTTGGCAAAGTTAATAACTTTGTTAAATTTGGCAGTGTATGTTTTTCGCTAAAGGATATACCCGGTGGTTGCGCAATGCCTATAGTGGCTAATTGGGCTTATAGTAAGTACTATACTATAGGCTCATTGTATGTTTCTTGGAGTGATGGTGATGGAATTACTGGCGTCCCACAACCTGTTGAGGGTGTTAGTTCCTTTATGAATGTTACATTGGACAAATGTACTAAATATAATATTTATGATGTATCTGGTGTGGGTGTTATTCGCGTTAGCAATGACACCTTTCTTAATGGAATTACGTACACATCAACTTCAGGTAACCTTCTGGGTTTTAAAGATGTTACTAAGGGCACCATCTACTCTATCACTCCTTGTAACCCACCAGATCAGCTTGTTGTTTATCAGCAAGCTGTTGTTGGTGCTATGTTGTCTGAAAATTTTACTAGTTACGGCTTTTCTAATGTTGTAGAACTGCCGAAATTTTTCTATGCGTCCAATGGCACTTATAATTGCACAGACGCTGTTTTAACTTATTCTAGTTTTGGCGTTTGTGCAGATGGTTCTATAATTGCTGTTCAACCACGTAATGTTTCATATGATAGTGTTTCAGCTATCGTCACAGCTAATTTGTCTATACCTTCCAATTGGACCACTTCGGTCCAGGTTGAGTATTTACAAATTACAAGTACACCTATCGTAGTTGATTGCTCCACTTATGTTTGCAATGGTAATGTGCGCTGTGTTGAATTGCTTAAGCAGTATACTTCTGCTTGTAAAACTATTGAAGACGCCTTAAGAAATAGCGCCAGGCTGGAGTCTGCAGATGTTAGTGAGATGCTCACTTTTGACAAGAAAGCGTTTACACTTGCTAATGTTAGTAGTTTTGGTGACTACAACCTTAGCAGCGTCATACCTAGCTTGCCCACAAGTGGTAGTAGAGTGGCTGGTCGCAGTGCCATAGAAGACATACTTTTTAGCAAACTTGTTACTTCTGGACTTGGCACTGTGGACGCAGACTACAAAAAGTGCACTAAGGGTCTTTCCATTGCTGACTTGGCTTGTGCTCAATATTATAATGGCATTATGGTTTTGCCTGGCGTCGCTGATGCTGAACGAATGGCCATGTATACAGGTTCTTTAATTGGTGGAATTGCTTTAGGAGGTCTAACATCAGCCGTTTCAATACCATTTTCATTAGCAATTCAGGCACGTTTAAATTATGTTGCATTGCAGACTGATGTTTTACAAGAAAATCAGAAAATTCTTGCTGCATCTTTTAACAAAGCAATGACCAACATAGTAGATGCCTTTACTGGTGTTAATGATGCTATTACACAAACTTCACAAGCCCTACAAACAGTTGCTACTGCACTTAACAAGATCCAGGATGTTGTTAATCAACAAGGCAACTCATTGAACCATTTAACTTCTCAGTTGAGGCAGAATTTTCAAGCTATCTCTAGCTCTATTCAGGCTATCTATGACAGACTTGACACTATTCAGGCTGATCAACAAGTAGATAGGCTGATTACTGGTAGATTGGCTGCTTTGAATGTATTCGTTTCTCATACATTGACTAAGTACACTGAAGTTCGTGCTTCCAGACAGCTTGCACAACAAAAAGTGAATGAGTGTGTCAAATCCCAGTCTAAGCGTTATGGCTTCTGTGGAAATGGCACTCACATTTTCTCAATTGTTAATGCTGCTCCTGAGGGGCTTGTTTTTCTCCACACTGTCTTGTTGCCGACACAATATAAGGATGTTGAAGCGTGGTCTGGGTTGTGCGTTGATGGTACAAACGGTTATGTGTTGCGACAACCTAATCTTGCTCTTTACAAAGAAGGCAATTATTATAGAATCACATCTCGCATAATGTTTGAACCACGTATTCCTACCATGGCAGATTTTGTTCAAATTGAAAATTGCAATGTCACATTTGTTAACATTTCTCGCTCTGAGTTGCAAACCATTGTGCCAGAGTATATTGATGTTAATAAGACGCTGCAAGAATTAAGTTACAAATTGCCAAATTACACTGTTCCAGACCTAGTTGTCGAACAGTACAACCAGACTATTTTGAATTTGACCAGTGAAATTAGCACCCTTGAAAATAAATCTGCGGAGCTTAATTACACTGTTCAAAAATTGCAAACTCTGATTGACAACATAAATAGCACATTAGTCGACTTAAAGTGGCTCAACCGGGTTGAGACTTACATCAAGTGGCCGTGGTGGGTGTGGTTGTGCATTTCAGTCGTGCTCATCTTTGTGGTGAGTATGTTGCTATTATGTTGTTGTTCTACTGGTTGCTGTGGCTTCTTTAGTTGTTTTGCATCTTCTATTAGAGGTTGTTGTGAATCAACTAAACTTCCTTATTACGACGTTGAAAAGATCCACATACAGTAA
Protein
MFVLLVAYALLHIAGCQTTNGLNTSYSVCNGCVGYSENVFAVESGGYIPSDFAFNNWFLLTNTSSVVDGVVRSFQPLLLNCLWSVSGLRFTTGFVYFNGTGRGDCKGFSSDVLSDVIRYNLNFEENLRRGTILFKTSYGVVVFYCTNNTLVSGDAHIPFGTVLGNFYCFVNTTIGNETTSAFVGALPKTVREFVISRTGHFYINGYRYFTLGNVEAVNFNVTTAETTDFCTVALASYADVLVNVSQTSIANIIYCNSVINRLRCDQLSFDVPDGFYSTSPIQSVELPVSIVSLPVYHKHTFIVLYVDFKPQSGGGKCFNCYPAGVNITLANFNETKGPLCVDTSHFTTKYVAVYANVGRWSASINTGNCPFSFGKVNNFVKFGSVCFSLKDIPGGCAMPIVANWAYSKYYTIGSLYVSWSDGDGITGVPQPVEGVSSFMNVTLDKCTKYNIYDVSGVGVIRVSNDTFLNGITYTSTSGNLLGFKDVTKGTIYSITPCNPPDQLVVYQQAVVGAMLSENFTSYGFSNVVELPKFFYASNGTYNCTDAVLTYSSFGVCADGSIIAVQPRNVSYDSVSAIVTANLSIPSNWTTSVQVEYLQITSTPIVVDCSTYVCNGNVRCVELLKQYTSACKTIEDALRNSARLESADVSEMLTFDKKAFTLANVSSFGDYNLSSVIPSLPTSGSRVAGRSAIEDILFSKLVTSGLGTVDADYKKCTKGLSIADLACAQYYNGIMVLPGVADAERMAMYTGSLIGGIALGGLTSAVSIPFSLAIQARLNYVALQTDVLQENQKILAASFNKAMTNIVDAFTGVNDAITQTSQALQTVATALNKIQDVVNQQGNSLNHLTSQLRQNFQAISSSIQAIYDRLDTIQADQQVDRLITGRLAALNVFVSHTLTKYTEVRASRQLAQQKVNECVKSQSKRYGFCGNGTHIFSIVNAAPEGLVFLHTVLLPTQYKDVEAWSGLCVDGTNGYVLRQPNLALYKEGNYYRITSRIMFEPRIPTMADFVQIENCNVTFVNISRSELQTIVPEYIDVNKTLQELSYKLPNYTVPDLVVEQYNQTILNLTSEISTLENKSAELNYTVQKLQTLIDNINSTLVDLKWLNRVETYIKWPWWVWLCISVVLIFVVSMLLLCCCSTGCCGFFSCFASSIRGCCESTKLPYYDVEKIHIQ
Summary
Function
S1 region attaches the virion to the cell membrane by interacting with host ANPEP/aminopeptidase N, initiating the infection. Binding to the receptor probably induces conformational changes in the S glycoprotein unmasking the fusion peptide of S2 region and activating membranes fusion. S2 region belongs to the class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) regions assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer. During virus morphogenesis, found in a complex with M and HE proteins. Interacts with host ANPEP.
Similarity
Belongs to the alphacoronaviruses spike protein family.
Uniprot
EMBL
ProteinModelPortal
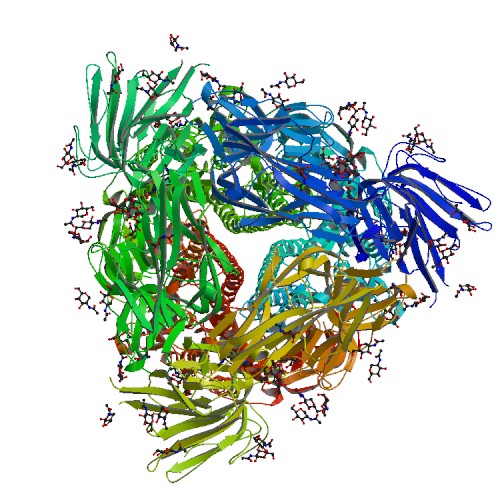
PDB
6U7H
E-value=0.0 Score=2211 Identity=99.10%
Cov(Q)=94.88% Cov(P)=96.03%
3D Structure
Ontologies
GO
GO:0019031 C:viral envelope
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0016021 C:integral component of membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0016021 C:integral component of membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
Subcellular Location
From MSLVP
Single-Pass Membrane
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
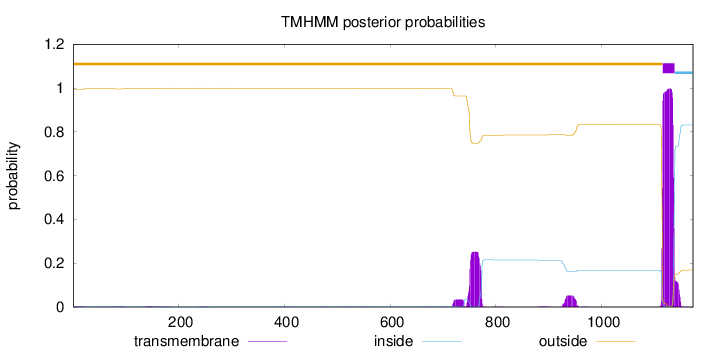
Topology
Length:
1173
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
31.31096
Exp number, first 60 AAs:
0.06343
Total prob of N-in:
0.00532
outside
1 - 1115
TMhelix
1116 - 1138
inside
1139 - 1173