Strain
CHINA_HS_2019_MN908947
(Region: China: Wuhan; Strain: Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, complete genome.; Date: 2019)
Gene
surface glycoprotein
Description
Annotated in NCBI,
surface glycoprotein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGTGTTAATCTTACAACCAGAACTCAATTACCCCCTGCATACACTAATTCTTTCACACGTGGTGTTTATTACCCTGACAAAGTTTTCAGATCCTCAGTTTTACATTCAACTCAGGACTTGTTCTTACCTTTCTTTTCCAATGTTACTTGGTTCCATGCTATACATGTCTCTGGGACCAATGGTACTAAGAGGTTTGATAACCCTGTCCTACCATTTAATGATGGTGTTTATTTTGCTTCCACTGAGAAGTCTAACATAATAAGAGGCTGGATTTTTGGTACTACTTTAGATTCGAAGACCCAGTCCCTACTTATTGTTAATAACGCTACTAATGTTGTTATTAAAGTCTGTGAATTTCAATTTTGTAATGATCCATTTTTGGGTGTTTATTACCACAAAAACAACAAAAGTTGGATGGAAAGTGAGTTCAGAGTTTATTCTAGTGCGAATAATTGCACTTTTGAATATGTCTCTCAGCCTTTTCTTATGGACCTTGAAGGAAAACAGGGTAATTTCAAAAATCTTAGGGAATTTGTGTTTAAGAATATTGATGGTTATTTTAAAATATATTCTAAGCACACGCCTATTAATTTAGTGCGTGATCTCCCTCAGGGTTTTTCGGCTTTAGAACCATTGGTAGATTTGCCAATAGGTATTAACATCACTAGGTTTCAAACTTTACTTGCTTTACATAGAAGTTATTTGACTCCTGGTGATTCTTCTTCAGGTTGGACAGCTGGTGCTGCAGCTTATTATGTGGGTTATCTTCAACCTAGGACTTTTCTATTAAAATATAATGAAAATGGAACCATTACAGATGCTGTAGACTGTGCACTTGACCCTCTCTCAGAAACAAAGTGTACGTTGAAATCCTTCACTGTAGAAAAAGGAATCTATCAAACTTCTAACTTTAGAGTCCAACCAACAGAATCTATTGTTAGATTTCCTAATATTACAAACTTGTGCCCTTTTGGTGAAGTTTTTAACGCCACCAGATTTGCATCTGTTTATGCTTGGAACAGGAAGAGAATCAGCAACTGTGTTGCTGATTATTCTGTCCTATATAATTCCGCATCATTTTCCACTTTTAAGTGTTATGGAGTGTCTCCTACTAAATTAAATGATCTCTGCTTTACTAATGTCTATGCAGATTCATTTGTAATTAGAGGTGATGAAGTCAGACAAATCGCTCCAGGGCAAACTGGAAAGATTGCTGATTATAATTATAAATTACCAGATGATTTTACAGGCTGCGTTATAGCTTGGAATTCTAACAATCTTGATTCTAAGGTTGGTGGTAATTATAATTACCTGTATAGATTGTTTAGGAAGTCTAATCTCAAACCTTTTGAGAGAGATATTTCAACTGAAATCTATCAGGCCGGTAGCACACCTTGTAATGGTGTTGAAGGTTTTAATTGTTACTTTCCTTTACAATCATATGGTTTCCAACCCACTAATGGTGTTGGTTACCAACCATACAGAGTAGTAGTACTTTCTTTTGAACTTCTACATGCACCAGCAACTGTTTGTGGACCTAAAAAGTCTACTAATTTGGTTAAAAACAAATGTGTCAATTTCAACTTCAATGGTTTAACAGGCACAGGTGTTCTTACTGAGTCTAACAAAAAGTTTCTGCCTTTCCAACAATTTGGCAGAGACATTGCTGACACTACTGATGCTGTCCGTGATCCACAGACACTTGAGATTCTTGACATTACACCATGTTCTTTTGGTGGTGTCAGTGTTATAACACCAGGAACAAATACTTCTAACCAGGTTGCTGTTCTTTATCAGGATGTTAACTGCACAGAAGTCCCTGTTGCTATTCATGCAGATCAACTTACTCCTACTTGGCGTGTTTATTCTACAGGTTCTAATGTTTTTCAAACACGTGCAGGCTGTTTAATAGGGGCTGAACATGTCAACAACTCATATGAGTGTGACATACCCATTGGTGCAGGTATATGCGCTAGTTATCAGACTCAGACTAATTCTCCTCGGCGGGCACGTAGTGTAGCTAGTCAATCCATCATTGCCTACACTATGTCACTTGGTGCAGAAAATTCAGTTGCTTACTCTAATAACTCTATTGCCATACCCACAAATTTTACTATTAGTGTTACCACAGAAATTCTACCAGTGTCTATGACCAAGACATCAGTAGATTGTACAATGTACATTTGTGGTGATTCAACTGAATGCAGCAATCTTTTGTTGCAATATGGCAGTTTTTGTACACAATTAAACCGTGCTTTAACTGGAATAGCTGTTGAACAAGACAAAAACACCCAAGAAGTTTTTGCACAAGTCAAACAAATTTACAAAACACCACCAATTAAAGATTTTGGTGGTTTTAATTTTTCACAAATATTACCAGATCCATCAAAACCAAGCAAGAGGTCATTTATTGAAGATCTACTTTTCAACAAAGTGACACTTGCAGATGCTGGCTTCATCAAACAATATGGTGATTGCCTTGGTGATATTGCTGCTAGAGACCTCATTTGTGCACAAAAGTTTAACGGCCTTACTGTTTTGCCACCTTTGCTCACAGATGAAATGATTGCTCAATACACTTCTGCACTGTTAGCGGGTACAATCACTTCTGGTTGGACCTTTGGTGCAGGTGCTGCATTACAAATACCATTTGCTATGCAAATGGCTTATAGGTTTAATGGTATTGGAGTTACACAGAATGTTCTCTATGAGAACCAAAAATTGATTGCCAACCAATTTAATAGTGCTATTGGCAAAATTCAAGACTCACTTTCTTCCACAGCAAGTGCACTTGGAAAACTTCAAGATGTGGTCAACCAAAATGCACAAGCTTTAAACACGCTTGTTAAACAACTTAGCTCCAATTTTGGTGCAATTTCAAGTGTTTTAAATGATATCCTTTCACGTCTTGACAAAGTTGAGGCTGAAGTGCAAATTGATAGGTTGATCACAGGCAGACTTCAAAGTTTGCAGACATATGTGACTCAACAATTAATTAGAGCTGCAGAAATCAGAGCTTCTGCTAATCTTGCTGCTACTAAAATGTCAGAGTGTGTACTTGGACAATCAAAAAGAGTTGATTTTTGTGGAAAGGGCTATCATCTTATGTCCTTCCCTCAGTCAGCACCTCATGGTGTAGTCTTCTTGCATGTGACTTATGTCCCTGCACAAGAAAAGAACTTCACAACTGCTCCTGCCATTTGTCATGATGGAAAAGCACACTTTCCTCGTGAAGGTGTCTTTGTTTCAAATGGCACACACTGGTTTGTAACACAAAGGAATTTTTATGAACCACAAATCATTACTACAGACAACACATTTGTGTCTGGTAACTGTGATGTTGTAATAGGAATTGTCAACAACACAGTTTATGATCCTTTGCAACCTGAATTAGACTCATTCAAGGAGGAGTTAGATAAATATTTTAAGAATCATACATCACCAGATGTTGATTTAGGTGACATCTCTGGCATTAATGCTTCAGTTGTAAACATTCAAAAAGAAATTGACCGCCTCAATGAGGTTGCCAAGAATTTAAATGAATCTCTCATCGATCTCCAAGAACTTGGAAAGTATGAGCAGTATATAAAATGGCCATGGTACATTTGGCTAGGTTTTATAGCTGGCTTGATTGCCATAGTAATGGTGACAATTATGCTTTGCTGTATGACCAGTTGCTGTAGTTGTCTCAAGGGCTGTTGTTCTTGTGGATCCTGCTGCAAATTTGATGAAGACGACTCTGAGCCAGTGCTCAAAGGAGTCAAATTACATTACACATAA
Protein
MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVKLHYT
Summary
Function
Spike protein S1: attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
attaches the virion to the cell membrane by interacting with host receptor, initiating the infection (By similarity). Binding to human ACE2 and CLEC4M/DC-SIGNR receptors and internalization of the virus into the endosomes of the host cell induces conformational changes in the S glycoprotein. Proteolysis by cathepsin CTSL may unmask the fusion peptide of S2 and activate membranes fusion within endosomes.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
attaches the virion to the cell membrane by interacting with host receptor, initiating the infection (By similarity). Binding to human ACE2 and CLEC4M/DC-SIGNR receptors and internalization of the virus into the endosomes of the host cell induces conformational changes in the S glycoprotein. Proteolysis by cathepsin CTSL may unmask the fusion peptide of S2 and activate membranes fusion within endosomes.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Subunit
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes.
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes (By similarity). Binds to human and palm civet ACE2 and human CLEC4M/DC-SIGNR. Interacts with the accessory proteins 3a and 7a.
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes (By similarity). Binds to human and palm civet ACE2 and human CLEC4M/DC-SIGNR. Interacts with the accessory proteins 3a and 7a.
Miscellaneous
Tor2 is the prototype of the virus isolated during the severe SARS outbreak in 2002-2003. GD03 has been isolated from the second mild SARS outbreak in winter 2003-2004. SZ3 has been isolated from palm civet, the presumed animal reservoir. The spike proteins from those three isolates display a strong affinity for palm civet ACE2 receptor, whereas only the Tor2 spike protein efficiently binds human ACE2. This may explain the high pathogenicity of Tor2 virus, whose spike is highly adapted to the human host. Therefore, the lack of severity of disease during the 2003-2004 outbreak could be due to the incomplete adaptation of GD03 virus to bind human ACE2. Mutation Asn-479 and Thr-487 in palm civet coronavirus seems necessary and sufficient for the virus to acquire the ability to efficiently infect humans.
Similarity
Belongs to the betacoronaviruses spike protein family.
Keywords
3D-structure
Coiled coil
Disulfide bond
Fusion of virus membrane with host endosomal membrane
Fusion of virus membrane with host membrane
Glycoprotein
Host cell membrane
Host membrane
Host-virus interaction
Lipoprotein
Membrane
Palmitate
Reference proteome
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Viral penetration into host cytoplasm
Virion
Virulence
Virus entry into host cell
Feature
chain Spike protein S1
Uniprot
A0A2R3SUW7
A0A2R3SUW9
A0A2D1PXA9
A0A2D1PX97
A0A0U2IWM2
A0A2D1PX29
+ More
U5WLK5 Q6TPE8 U5WHZ7 U5WI05 A0A2D1PXC0 A0A4Y6GL47 A0A2D1PXD5 Q5GDB6 Q6DSU4 Q4JDM4 Q6GYR1 Q1EFP5 Q1EFN2 Q5DIC5 Q5GDB4 D2E206 A4ZF26 D2E1K6 Q6T7X5 Q3ZTC8 A4ZF28 Q202F5 Q202F4 Q5GDB3 Q4JDN5 Q4JDN2 Q3ZTC5 Q3ZTE0 D2E250 Q4JDM7 Q5GDJ7 Q4JDM9 Q4JDN0 Q4JDN3 Q202G5 Q202G3 Q202G8 A4ZF27 Q3ZTC7 Q4JDM6 Q6RD20 Q4JDN1 Q6RD64 Q6VAA0 Q6UZF4 Q6VA78 Q6UZF0 Q6RCY7 Q6RD31 Q6RD09 Q202H5 Q6JH38 Q6RCX6 Q3S2D2 Q6RD42 Q6VA89 Q6RCZ8 Q6RCW5 Q6RD53 Q6WGP3 Q6V585 Q202E9 A7J8L4 Q309G9 A0A3G5BJ39 D2E1E7 D2E1D2 D3KDQ3 P59594 Q202E6 Q4JDN4 Q5Y187 Q6T7Y1 Q5GDB5 D2E2T4 D2E2D8 Q6R7Y6 A4ZF30 Q5GDB7 Q4JDP0 Q6JH46 Q4JDP1 Q5GDJ6 D2E1Z1 D2E2M5 D2E1H7 Q3ZTF3 Q202F2 Q4JDN9 Q4JDM5 A4ZF29 Q4JDN8 Q4JDM3 Q692E4
U5WLK5 Q6TPE8 U5WHZ7 U5WI05 A0A2D1PXC0 A0A4Y6GL47 A0A2D1PXD5 Q5GDB6 Q6DSU4 Q4JDM4 Q6GYR1 Q1EFP5 Q1EFN2 Q5DIC5 Q5GDB4 D2E206 A4ZF26 D2E1K6 Q6T7X5 Q3ZTC8 A4ZF28 Q202F5 Q202F4 Q5GDB3 Q4JDN5 Q4JDN2 Q3ZTC5 Q3ZTE0 D2E250 Q4JDM7 Q5GDJ7 Q4JDM9 Q4JDN0 Q4JDN3 Q202G5 Q202G3 Q202G8 A4ZF27 Q3ZTC7 Q4JDM6 Q6RD20 Q4JDN1 Q6RD64 Q6VAA0 Q6UZF4 Q6VA78 Q6UZF0 Q6RCY7 Q6RD31 Q6RD09 Q202H5 Q6JH38 Q6RCX6 Q3S2D2 Q6RD42 Q6VA89 Q6RCZ8 Q6RCW5 Q6RD53 Q6WGP3 Q6V585 Q202E9 A7J8L4 Q309G9 A0A3G5BJ39 D2E1E7 D2E1D2 D3KDQ3 P59594 Q202E6 Q4JDN4 Q5Y187 Q6T7Y1 Q5GDB5 D2E2T4 D2E2D8 Q6R7Y6 A4ZF30 Q5GDB7 Q4JDP0 Q6JH46 Q4JDP1 Q5GDJ6 D2E1Z1 D2E2M5 D2E1H7 Q3ZTF3 Q202F2 Q4JDN9 Q4JDM5 A4ZF29 Q4JDN8 Q4JDM3 Q692E4
Pubmed
29190287
26719272
24172901
15695582
16140765
16625834
+ More
18987164 14569023 20463816 16485471 17112780 14983045 14654320 16269206 14527350 14733771 16112670 19370068 17184168 30393776 22412934 12730500 12730501 12853594 12876307 12958366 12781537 12917450 14647384 14670965 15518555 15450134 15496474 15313178 15890958 15831954 15791205 16081529 15194747 16840309 16519916 17134730 17166901 19321428 20861307 15345712 15604146 14511651 16166518 16698550 15757562
18987164 14569023 20463816 16485471 17112780 14983045 14654320 16269206 14527350 14733771 16112670 19370068 17184168 30393776 22412934 12730500 12730501 12853594 12876307 12958366 12781537 12917450 14647384 14670965 15518555 15450134 15496474 15313178 15890958 15831954 15791205 16081529 15194747 16840309 16519916 17134730 17166901 19321428 20861307 15345712 15604146 14511651 16166518 16698550 15757562
EMBL
MG772933
AVP78031.1
MG772934
AVP78042.1
KY417146
ATO98157.1
+ More
KY417150 ATO98205.1 KT444582 ALK02457.1 KY417144 ATO98132.1 KC881005 AGZ48806.1 AY390556 AAS00003.1 KC881006 AGZ48818.1 KC881007 KF367457 AGZ48828.1 AGZ48831.1 KY417151 ATO98218.1 MK211376 QDF43825.1 KY417152 ATO98231.1 AY627045 AAV49720.1 AY648300 AAT74874.1 AY687372 AAV98002.1 AY595412 AAT52330.1 AY864805 AAY60780.1 AY864806 AAY60793.1 AY772062 AAX16192.1 AY627047 AAV49722.1 FJ882941 ACZ71976.1 DQ514528 ABF68955.1 FJ882931 FJ882954 FJ882955 ACZ71826.1 ACZ72166.1 AY429078 AAR07630.1 AY572036 AAU04661.1 DQ514530 ABF68957.1 DQ412585 DQ412614 ABD72977.1 ABD72992.1 DQ412597 DQ412598 DQ412612 DQ412615 ABD72985.1 ABD72986.1 ABD72990.1 ABD72993.1 AY627048 AAV49723.1 AY687361 AAV97991.1 AY687364 AAV97994.1 AY572038 AAU04664.1 AY572035 AAU04649.1 FJ882944 ACZ72020.1 AY687369 AAV97999.1 AY613951 AAU93318.1 AY687367 AAV97997.1 AY687366 AAV97996.1 AY686864 AY687363 AAV49730.1 AAV97993.1 DQ412594 ABD72982.1 DQ412596 ABD72984.1 DQ412588 ABD72979.1 DQ514529 ABF68956.1 AY572037 AAU04662.1 AY687370 AAV98000.1 AY502927 AAR87545.1 AY687365 AAV97995.1 AY502923 AAR87501.1 AY345986 AAP94737.1 AY357075 AAR14807.1 AY345988 AAP94759.1 AY357076 AAR14811.1 AY502930 AAR87578.1 AY502926 AAR87534.1 AY502928 AAR87556.1 DQ412578 ABD72972.1 AY485278 AAR23258.1 AY502931 AAR87589.1 DQ182595 ABA02260.1 AY502925 AAR87523.1 AY345987 AAP94748.1 AY502929 AAR87567.1 AY502932 AAR87600.1 AY502924 AAR87512.1 AY286320 AAR33050.1 AY350750 AAR14803.1 DQ412574 DQ412581 DQ412587 DQ412589 DQ412590 DQ412595 DQ412605 DQ412611 DQ412613 DQ412616 DQ412618 DQ412624 ABD72968.1 ABD72975.1 ABD72978.1 ABD72980.1 ABD72981.1 ABD72983.1 ABD72987.1 ABD72989.1 ABD72991.1 ABD72994.1 ABD72996.1 ABD72998.1 DQ898174 JX163924 JX163928 ABI96958.1 AFR58686.1 AFR58742.1 DQ231462 EU371559 ABB29898.2 ACB69849.1 MK062179 MK062181 MK062183 AYV99803.1 FJ882927 FJ882932 FJ882933 FJ882934 FJ882935 FJ882936 FJ882937 FJ882938 FJ882939 FJ882946 FJ882947 JF292921 KF514388 KF514392 KF514394 KF514396 KF514397 KF514398 KF514399 KF514400 KF514404 KF514408 KF514409 KF514413 KF514415 KF514418 KF514419 KF514421 KF514422 KF514423 ACZ71767.1 ACZ71841.1 AEA10998.1 AGT20793.1 AGT20853.1 AGT20883.1 AGT20913.1 AGT20928.1 AGT20943.1 AGT20958.1 AGT20973.1 AGT21033.1 AGT21093.1 AGT21108.1 AGT21168.1 AGT21198.1 AGT21243.1 AGT21258.1 AGT21288.1 AGT21303.1 AGT21318.1 FJ882926 FJ882928 FJ882930 JF292922 KF514389 KF514390 KF514391 KF514393 KF514395 KF514401 KF514402 KF514403 KF514405 KF514406 KF514410 KF514411 KF514412 KF514414 KF514416 KF514417 KF514420 ACZ71752.1 AEA11013.1 AGT20808.1 AGT20823.1 AGT20838.1 AGT20868.1 AGT20898.1 AGT20988.1 AGT21003.1 AGT21018.1 AGT21048.1 AGT21063.1 AGT21123.1 AGT21138.1 AGT21153.1 AGT21183.1 AGT21213.1 AGT21228.1 AGT21273.1 GU553365 JQ316196 JN854286 ADC35511.1 AFD56973.1 AFK10254.1 AY278741 AY274119 AY282752 AY278554 AY278491 AY304495 AY304492 AY278487 AY278488 AY278490 AY279354 AY278489 AY283794 AY283795 AY283796 AY283797 AY283798 AY291451 AY310120 AY291315 AY304486 AY321118 AY323976 AY322207 AY338174 AY338175 AY348314 AP006557 AP006558 AP006559 AP006560 AP006561 AY323977 AY362698 AY362699 AY427439 AY463059 AY525636 DQ412628 ABD73001.1 AY687362 AAV97992.1 AY714217 AAU81608.1 AY429072 AAR07624.1 AY627046 AAV49721.1 FJ882960 ACZ72254.1 FJ882950 ACZ72108.1 AY508724 AAR91586.1 DQ514532 ABF68959.1 AY627044 AAV49719.1 AY687356 AAV97986.1 AY485277 AAR23250.1 AY687355 AAV97985.1 AY613952 AAU93319.1 FJ882940 ACZ71961.1 FJ882956 ACZ72195.1 FJ882929 ACZ71797.1 AY572034 AAU04646.1 DQ412617 ABD72995.1 AY687357 AAV97987.1 AY687371 AAV98001.1 DQ514531 ABF68958.1 AY687358 AAV97988.1 AY686863 AY687373 AAV91631.1 AAV98003.1 AY654624 AAT76147.1
KY417150 ATO98205.1 KT444582 ALK02457.1 KY417144 ATO98132.1 KC881005 AGZ48806.1 AY390556 AAS00003.1 KC881006 AGZ48818.1 KC881007 KF367457 AGZ48828.1 AGZ48831.1 KY417151 ATO98218.1 MK211376 QDF43825.1 KY417152 ATO98231.1 AY627045 AAV49720.1 AY648300 AAT74874.1 AY687372 AAV98002.1 AY595412 AAT52330.1 AY864805 AAY60780.1 AY864806 AAY60793.1 AY772062 AAX16192.1 AY627047 AAV49722.1 FJ882941 ACZ71976.1 DQ514528 ABF68955.1 FJ882931 FJ882954 FJ882955 ACZ71826.1 ACZ72166.1 AY429078 AAR07630.1 AY572036 AAU04661.1 DQ514530 ABF68957.1 DQ412585 DQ412614 ABD72977.1 ABD72992.1 DQ412597 DQ412598 DQ412612 DQ412615 ABD72985.1 ABD72986.1 ABD72990.1 ABD72993.1 AY627048 AAV49723.1 AY687361 AAV97991.1 AY687364 AAV97994.1 AY572038 AAU04664.1 AY572035 AAU04649.1 FJ882944 ACZ72020.1 AY687369 AAV97999.1 AY613951 AAU93318.1 AY687367 AAV97997.1 AY687366 AAV97996.1 AY686864 AY687363 AAV49730.1 AAV97993.1 DQ412594 ABD72982.1 DQ412596 ABD72984.1 DQ412588 ABD72979.1 DQ514529 ABF68956.1 AY572037 AAU04662.1 AY687370 AAV98000.1 AY502927 AAR87545.1 AY687365 AAV97995.1 AY502923 AAR87501.1 AY345986 AAP94737.1 AY357075 AAR14807.1 AY345988 AAP94759.1 AY357076 AAR14811.1 AY502930 AAR87578.1 AY502926 AAR87534.1 AY502928 AAR87556.1 DQ412578 ABD72972.1 AY485278 AAR23258.1 AY502931 AAR87589.1 DQ182595 ABA02260.1 AY502925 AAR87523.1 AY345987 AAP94748.1 AY502929 AAR87567.1 AY502932 AAR87600.1 AY502924 AAR87512.1 AY286320 AAR33050.1 AY350750 AAR14803.1 DQ412574 DQ412581 DQ412587 DQ412589 DQ412590 DQ412595 DQ412605 DQ412611 DQ412613 DQ412616 DQ412618 DQ412624 ABD72968.1 ABD72975.1 ABD72978.1 ABD72980.1 ABD72981.1 ABD72983.1 ABD72987.1 ABD72989.1 ABD72991.1 ABD72994.1 ABD72996.1 ABD72998.1 DQ898174 JX163924 JX163928 ABI96958.1 AFR58686.1 AFR58742.1 DQ231462 EU371559 ABB29898.2 ACB69849.1 MK062179 MK062181 MK062183 AYV99803.1 FJ882927 FJ882932 FJ882933 FJ882934 FJ882935 FJ882936 FJ882937 FJ882938 FJ882939 FJ882946 FJ882947 JF292921 KF514388 KF514392 KF514394 KF514396 KF514397 KF514398 KF514399 KF514400 KF514404 KF514408 KF514409 KF514413 KF514415 KF514418 KF514419 KF514421 KF514422 KF514423 ACZ71767.1 ACZ71841.1 AEA10998.1 AGT20793.1 AGT20853.1 AGT20883.1 AGT20913.1 AGT20928.1 AGT20943.1 AGT20958.1 AGT20973.1 AGT21033.1 AGT21093.1 AGT21108.1 AGT21168.1 AGT21198.1 AGT21243.1 AGT21258.1 AGT21288.1 AGT21303.1 AGT21318.1 FJ882926 FJ882928 FJ882930 JF292922 KF514389 KF514390 KF514391 KF514393 KF514395 KF514401 KF514402 KF514403 KF514405 KF514406 KF514410 KF514411 KF514412 KF514414 KF514416 KF514417 KF514420 ACZ71752.1 AEA11013.1 AGT20808.1 AGT20823.1 AGT20838.1 AGT20868.1 AGT20898.1 AGT20988.1 AGT21003.1 AGT21018.1 AGT21048.1 AGT21063.1 AGT21123.1 AGT21138.1 AGT21153.1 AGT21183.1 AGT21213.1 AGT21228.1 AGT21273.1 GU553365 JQ316196 JN854286 ADC35511.1 AFD56973.1 AFK10254.1 AY278741 AY274119 AY282752 AY278554 AY278491 AY304495 AY304492 AY278487 AY278488 AY278490 AY279354 AY278489 AY283794 AY283795 AY283796 AY283797 AY283798 AY291451 AY310120 AY291315 AY304486 AY321118 AY323976 AY322207 AY338174 AY338175 AY348314 AP006557 AP006558 AP006559 AP006560 AP006561 AY323977 AY362698 AY362699 AY427439 AY463059 AY525636 DQ412628 ABD73001.1 AY687362 AAV97992.1 AY714217 AAU81608.1 AY429072 AAR07624.1 AY627046 AAV49721.1 FJ882960 ACZ72254.1 FJ882950 ACZ72108.1 AY508724 AAR91586.1 DQ514532 ABF68959.1 AY627044 AAV49719.1 AY687356 AAV97986.1 AY485277 AAR23250.1 AY687355 AAV97985.1 AY613952 AAU93319.1 FJ882940 ACZ71961.1 FJ882956 ACZ72195.1 FJ882929 ACZ71797.1 AY572034 AAU04646.1 DQ412617 ABD72995.1 AY687357 AAV97987.1 AY687371 AAV98001.1 DQ514531 ABF68958.1 AY687358 AAV97988.1 AY686863 AY687373 AAV91631.1 AAV98003.1 AY654624 AAT76147.1
Proteomes
UP000100289
UP000138593
UP000166164
UP000157984
UP000110240
UP000158790
+ More
UP000133349 UP000149347 UP000156013 UP000180369 UP000180367 UP000180372 UP000180373 UP000153024 UP000141980 UP000180370 UP000126514 UP000174857 UP000148533 UP000133546 UP000118579 UP000115935 UP000147828 UP000125082 UP000148329 UP000124386 UP000168969 UP000132037 UP000163176 UP000139901 UP000129608 UP000099158 UP000110133 UP000166558 UP000170944 UP000135071 UP000166757 UP000180361 UP000099142 UP000097931 UP000121467 UP000125925 UP000128288 UP000150617 UP000151673 UP000151756 UP000153830 UP000155701 UP000157930 UP000161442 UP000161729 UP000162782 UP000164349 UP000169779 UP000173963 UP000174280 UP000180434 UP000180435 UP000180436 UP000180437 UP000180438 UP000180439 UP000180440 UP000180441 UP000180442 UP000180443 UP000180444 UP000180445 UP000180446 UP000097852 UP000106870 UP000117796 UP000119316 UP000119820 UP000124542 UP000125968 UP000130610 UP000133348 UP000141212 UP000143912 UP000145152 UP000149516 UP000166143 UP000171773 UP000180363 UP000180364 UP000180366 UP000180376 UP000180377 UP000180378 UP000112510 UP000152329 UP000180382 UP000000354 UP000103670 UP000109640 UP000116947 UP000121636 UP000131569 UP000131955 UP000137377 UP000138690 UP000143093 UP000145651 UP000146108 UP000146181 UP000146296 UP000148194 UP000153467 UP000160648 UP000164441 UP000172416 UP000171372 UP000180375 UP000180371 UP000117876 UP000123303 UP000180368 UP000180374 UP000180365 UP000127950 UP000115496 UP000101079
UP000133349 UP000149347 UP000156013 UP000180369 UP000180367 UP000180372 UP000180373 UP000153024 UP000141980 UP000180370 UP000126514 UP000174857 UP000148533 UP000133546 UP000118579 UP000115935 UP000147828 UP000125082 UP000148329 UP000124386 UP000168969 UP000132037 UP000163176 UP000139901 UP000129608 UP000099158 UP000110133 UP000166558 UP000170944 UP000135071 UP000166757 UP000180361 UP000099142 UP000097931 UP000121467 UP000125925 UP000128288 UP000150617 UP000151673 UP000151756 UP000153830 UP000155701 UP000157930 UP000161442 UP000161729 UP000162782 UP000164349 UP000169779 UP000173963 UP000174280 UP000180434 UP000180435 UP000180436 UP000180437 UP000180438 UP000180439 UP000180440 UP000180441 UP000180442 UP000180443 UP000180444 UP000180445 UP000180446 UP000097852 UP000106870 UP000117796 UP000119316 UP000119820 UP000124542 UP000125968 UP000130610 UP000133348 UP000141212 UP000143912 UP000145152 UP000149516 UP000166143 UP000171773 UP000180363 UP000180364 UP000180366 UP000180376 UP000180377 UP000180378 UP000112510 UP000152329 UP000180382 UP000000354 UP000103670 UP000109640 UP000116947 UP000121636 UP000131569 UP000131955 UP000137377 UP000138690 UP000143093 UP000145651 UP000146108 UP000146181 UP000146296 UP000148194 UP000153467 UP000160648 UP000164441 UP000172416 UP000171372 UP000180375 UP000180371 UP000117876 UP000123303 UP000180368 UP000180374 UP000180365 UP000127950 UP000115496 UP000101079
PRIDE
Interpro
SUPFAM
SSF143587
SSF143587
Gene 3D
ProteinModelPortal
A0A2R3SUW7
A0A2R3SUW9
A0A2D1PXA9
A0A2D1PX97
A0A0U2IWM2
A0A2D1PX29
+ More
U5WLK5 Q6TPE8 U5WHZ7 U5WI05 A0A2D1PXC0 A0A4Y6GL47 A0A2D1PXD5 Q5GDB6 Q6DSU4 Q4JDM4 Q6GYR1 Q1EFP5 Q1EFN2 Q5DIC5 Q5GDB4 D2E206 A4ZF26 D2E1K6 Q6T7X5 Q3ZTC8 A4ZF28 Q202F5 Q202F4 Q5GDB3 Q4JDN5 Q4JDN2 Q3ZTC5 Q3ZTE0 D2E250 Q4JDM7 Q5GDJ7 Q4JDM9 Q4JDN0 Q4JDN3 Q202G5 Q202G3 Q202G8 A4ZF27 Q3ZTC7 Q4JDM6 Q6RD20 Q4JDN1 Q6RD64 Q6VAA0 Q6UZF4 Q6VA78 Q6UZF0 Q6RCY7 Q6RD31 Q6RD09 Q202H5 Q6JH38 Q6RCX6 Q3S2D2 Q6RD42 Q6VA89 Q6RCZ8 Q6RCW5 Q6RD53 Q6WGP3 Q6V585 Q202E9 A7J8L4 Q309G9 A0A3G5BJ39 D2E1E7 D2E1D2 D3KDQ3 P59594 Q202E6 Q4JDN4 Q5Y187 Q6T7Y1 Q5GDB5 D2E2T4 D2E2D8 Q6R7Y6 A4ZF30 Q5GDB7 Q4JDP0 Q6JH46 Q4JDP1 Q5GDJ6 D2E1Z1 D2E2M5 D2E1H7 Q3ZTF3 Q202F2 Q4JDN9 Q4JDM5 A4ZF29 Q4JDN8 Q4JDM3 Q692E4
U5WLK5 Q6TPE8 U5WHZ7 U5WI05 A0A2D1PXC0 A0A4Y6GL47 A0A2D1PXD5 Q5GDB6 Q6DSU4 Q4JDM4 Q6GYR1 Q1EFP5 Q1EFN2 Q5DIC5 Q5GDB4 D2E206 A4ZF26 D2E1K6 Q6T7X5 Q3ZTC8 A4ZF28 Q202F5 Q202F4 Q5GDB3 Q4JDN5 Q4JDN2 Q3ZTC5 Q3ZTE0 D2E250 Q4JDM7 Q5GDJ7 Q4JDM9 Q4JDN0 Q4JDN3 Q202G5 Q202G3 Q202G8 A4ZF27 Q3ZTC7 Q4JDM6 Q6RD20 Q4JDN1 Q6RD64 Q6VAA0 Q6UZF4 Q6VA78 Q6UZF0 Q6RCY7 Q6RD31 Q6RD09 Q202H5 Q6JH38 Q6RCX6 Q3S2D2 Q6RD42 Q6VA89 Q6RCZ8 Q6RCW5 Q6RD53 Q6WGP3 Q6V585 Q202E9 A7J8L4 Q309G9 A0A3G5BJ39 D2E1E7 D2E1D2 D3KDQ3 P59594 Q202E6 Q4JDN4 Q5Y187 Q6T7Y1 Q5GDB5 D2E2T4 D2E2D8 Q6R7Y6 A4ZF30 Q5GDB7 Q4JDP0 Q6JH46 Q4JDP1 Q5GDJ6 D2E1Z1 D2E2M5 D2E1H7 Q3ZTF3 Q202F2 Q4JDN9 Q4JDM5 A4ZF29 Q4JDN8 Q4JDM3 Q692E4
PDB
6ACK
E-value=0,
Score=4890
Ontologies
KEGG
GO
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0061025 P:membrane fusion
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0020002 C:host cell plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0042802 F:identical protein binding
GO:0046789 F:host cell surface receptor binding
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0061025 P:membrane fusion
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0020002 C:host cell plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0042802 F:identical protein binding
GO:0046789 F:host cell surface receptor binding
Subcellular Location
From MSLVP
Host nucleus > host nucleolus. Host cytoplasm. Secreted.
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
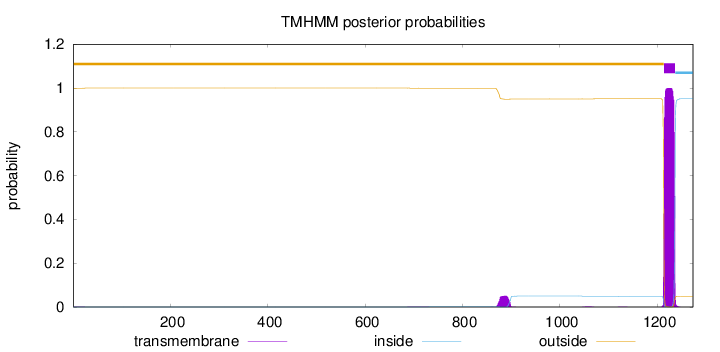
Topology
Length:
1273
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.97303
Exp number, first 60 AAs:
0.01558
Total prob of N-in:
0.00077
outside
1 - 1213
TMhelix
1214 - 1236
inside
1237 - 1273
Population Genetic Test Statistics
Pi
0.5507653
Theta
0.7009139
Tajima's D
-1.2445258
CLR
4.828786
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 |
Protein |
QHD43416.1 |
|
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 |
Protein |
NP_828851.1 |
|
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 |
Protein |
YP_003858584.1 |
|
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.