Strain
CANADA_HS_2003_NC_004718
(Region: Canada: Toronto; Strain: SARS coronavirus, complete genome.; Date: 2003)
Gene
orf1ab polyprotein (pp1ab)
Description
Annotated in NCBI,
orf1ab polyprotein (pp1ab)
Location
GenBank Accession
Full name
Replicase polyprotein 1ab
Alternative Name
ORF1ab polyprotein
Sequence
CDS
ATGGAGAGCCTTGTTCTTGGTGTCAACGAGAAAACACACGTCCAACTCAGTTTGCCTGTCCTTCAGGTTAGAGACGTGCTAGTGCGTGGCTTCGGGGACTCTGTGGAAGAGGCCCTATCGGAGGCACGTGAACACCTCAAAAATGGCACTTGTGGTCTAGTAGAGCTGGAAAAAGGCGTACTGCCCCAGCTTGAACAGCCCTATGTGTTCATTAAACGTTCTGATGCCTTAAGCACCAATCACGGCCACAAGGTCGTTGAGCTGGTTGCAGAAATGGACGGCATTCAGTACGGTCGTAGCGGTATAACACTGGGAGTACTCGTGCCACATGTGGGCGAAACCCCAATTGCATACCGCAATGTTCTTCTTCGTAAGAACGGTAATAAGGGAGCCGGTGGTCATAGCTATGGCATCGATCTAAAGTCTTATGACTTAGGTGACGAGCTTGGCACTGATCCCATTGAAGATTATGAACAAAACTGGAACACTAAGCATGGCAGTGGTGCACTCCGTGAACTCACTCGTGAGCTCAATGGAGGTGCAGTCACTCGCTATGTCGACAACAATTTCTGTGGCCCAGATGGGTACCCTCTTGATTGCATCAAAGATTTTCTCGCACGCGCGGGCAAGTCAATGTGCACTCTTTCCGAACAACTTGATTACATCGAGTCGAAGAGAGGTGTCTACTGCTGCCGTGACCATGAGCATGAAATTGCCTGGTTCACTGAGCGCTCTGATAAGAGCTACGAGCACCAGACACCCTTCGAAATTAAGAGTGCCAAGAAATTTGACACTTTCAAAGGGGAATGCCCAAAGTTTGTGTTTCCTCTTAACTCAAAAGTCAAAGTCATTCAACCACGTGTTGAAAAGAAAAAGACTGAGGGTTTCATGGGGCGTATACGCTCTGTGTACCCTGTTGCATCTCCACAGGAGTGTAACAATATGCACTTGTCTACCTTGATGAAATGTAATCATTGCGATGAAGTTTCATGGCAGACGTGCGACTTTCTGAAAGCCACTTGTGAACATTGTGGCACTGAAAATTTAGTTATTGAAGGACCTACTACATGTGGGTACCTACCTACTAATGCTGTAGTGAAAATGCCATGTCCTGCCTGTCAAGACCCAGAGATTGGACCTGAGCATAGTGTTGCAGATTATCACAACCACTCAAACATTGAAACTCGACTCCGCAAGGGAGGTAGGACTAGATGTTTTGGAGGCTGTGTGTTTGCCTATGTTGGCTGCTATAATAAGCGTGCCTACTGGGTTCCTCGTGCTAGTGCTGATATTGGCTCAGGCCATACTGGCATTACTGGTGACAATGTGGAGACCTTGAATGAGGATCTCCTTGAGATACTGAGTCGTGAACGTGTTAACATTAACATTGTTGGCGATTTTCATTTGAATGAAGAGGTTGCCATCATTTTGGCATCTTTCTCTGCTTCTACAAGTGCCTTTATTGACACTATAAAGAGTCTTGATTACAAGTCTTTCAAAACCATTGTTGAGTCCTGCGGTAACTATAAAGTTACCAAGGGAAAGCCCGTAAAAGGTGCTTGGAACATTGGACAACAGAGATCAGTTTTAACACCACTGTGTGGTTTTCCCTCACAGGCTGCTGGTGTTATCAGATCAATTTTTGCGCGCACACTTGATGCAGCAAACCACTCAATTCCTGATTTGCAAAGAGCAGCTGTCACCATACTTGATGGTATTTCTGAACAGTCATTACGTCTTGTCGACGCCATGGTTTATACTTCAGACCTGCTCACCAACAGTGTCATTATTATGGCATATGTAACTGGTGGTCTTGTACAACAGACTTCTCAGTGGTTGTCTAATCTTTTGGGCACTACTGTTGAAAAACTCAGGCCTATCTTTGAATGGATTGAGGCGAAACTTAGTGCAGGAGTTGAATTTCTCAAGGATGCTTGGGAGATTCTCAAATTTCTCATTACAGGTGTTTTTGACATCGTCAAGGGTCAAATACAGGTTGCTTCAGATAACATCAAGGATTGTGTAAAATGCTTCATTGATGTTGTTAACAAGGCACTCGAAATGTGCATTGATCAAGTCACTATCGCTGGCGCAAAGTTGCGATCACTCAACTTAGGTGAAGTCTTCATCGCTCAAAGCAAGGGACTTTACCGTCAGTGTATACGTGGCAAGGAGCAGCTGCAACTACTCATGCCTCTTAAGGCACCAAAAGAAGTAACCTTTCTTGAAGGTGATTCACATGACACAGTACTTACCTCTGAGGAGGTTGTTCTCAAGAACGGTGAACTCGAAGCACTCGAGACGCCCGTTGATAGCTTCACAAATGGAGCTATCGTTGGCACACCAGTCTGTGTAAATGGCCTCATGCTCTTAGAGATTAAGGACAAAGAACAATACTGCGCATTGTCTCCTGGTTTACTGGCTACAAACAATGTCTTTCGCTTAAAAGGGGGTGCACCAATTAAAGGTGTAACCTTTGGAGAAGATACTGTTTGGGAAGTTCAAGGTTACAAGAATGTGAGAATCACATTTGAGCTTGATGAACGTGTTGACAAAGTGCTTAATGAAAAGTGCTCTGTCTACACTGTTGAATCCGGTACCGAAGTTACTGAGTTTGCATGTGTTGTAGCAGAGGCTGTTGTGAAGACTTTACAACCAGTTTCTGATCTCCTTACCAACATGGGTATTGATCTTGATGAGTGGAGTGTAGCTACATTCTACTTATTTGATGATGCTGGTGAAGAAAACTTTTCATCACGTATGTATTGTTCCTTTTACCCTCCAGATGAGGAAGAAGAGGACGATGCAGAGTGTGAGGAAGAAGAAATTGATGAAACCTGTGAACATGAGTACGGTACAGAGGATGATTATCAAGGTCTCCCTCTGGAATTTGGTGCCTCAGCTGAAACAGTTCGAGTTGAGGAAGAAGAAGAGGAAGACTGGCTGGATGATACTACTGAGCAATCAGAGATTGAGCCAGAACCAGAACCTACACCTGAAGAACCAGTTAATCAGTTTACTGGTTATTTAAAACTTACTGACAATGTTGCCATTAAATGTGTTGACATCGTTAAGGAGGCACAAAGTGCTAATCCTATGGTGATTGTAAATGCTGCTAACATACACCTGAAACATGGTGGTGGTGTAGCAGGTGCACTCAACAAGGCAACCAATGGTGCCATGCAAAAGGAGAGTGATGATTACATTAAGCTAAATGGCCCTCTTACAGTAGGAGGGTCTTGTTTGCTTTCTGGACATAATCTTGCTAAGAAGTGTCTGCATGTTGTTGGACCTAACCTAAATGCAGGTGAGGACATCCAGCTTCTTAAGGCAGCATATGAAAATTTCAATTCACAGGACATCTTACTTGCACCATTGTTGTCAGCAGGCATATTTGGTGCTAAACCACTTCAGTCTTTACAAGTGTGCGTGCAGACGGTTCGTACACAGGTTTATATTGCAGTCAATGACAAAGCTCTTTATGAGCAGGTTGTCATGGATTATCTTGATAACCTGAAGCCTAGAGTGGAAGCACCTAAACAAGAGGAGCCACCAAACACAGAAGATTCCAAAACTGAGGAGAAATCTGTCGTACAGAAGCCTGTCGATGTGAAGCCAAAAATTAAGGCCTGCATTGATGAGGTTACCACAACACTGGAAGAAACTAAGTTTCTTACCAATAAGTTACTCTTGTTTGCTGATATCAATGGTAAGCTTTACCATGATTCTCAGAACATGCTTAGAGGTGAAGATATGTCTTTCCTTGAGAAGGATGCACCTTACATGGTAGGTGATGTTATCACTAGTGGTGATATCACTTGTGTTGTAATACCCTCCAAAAAGGCTGGTGGCACTACTGAGATGCTCTCAAGAGCTTTGAAGAAAGTGCCAGTTGATGAGTATATAACCACGTACCCTGGACAAGGATGTGCTGGTTATACACTTGAGGAAGCTAAGACTGCTCTTAAGAAATGCAAATCTGCATTTTATGTACTACCTTCAGAAGCACCTAATGCTAAGGAAGAGATTCTAGGAACTGTATCCTGGAATTTGAGAGAAATGCTTGCTCATGCTGAAGAGACAAGAAAATTAATGCCTATATGCATGGATGTTAGAGCCATAATGGCAACCATCCAACGTAAGTATAAAGGAATTAAAATTCAAGAGGGCATCGTTGACTATGGTGTCCGATTCTTCTTTTATACTAGTAAAGAGCCTGTAGCTTCTATTATTACGAAGCTGAACTCTCTAAATGAGCCGCTTGTCACAATGCCAATTGGTTATGTGACACATGGTTTTAATCTTGAAGAGGCTGCGCGCTGTATGCGTTCTCTTAAAGCTCCTGCCGTAGTGTCAGTATCATCACCAGATGCTGTTACTACATATAATGGATACCTCACTTCGTCATCAAAGACATCTGAGGAGCACTTTGTAGAAACAGTTTCTTTGGCTGGCTCTTACAGAGATTGGTCCTATTCAGGACAGCGTACAGAGTTAGGTGTTGAATTTCTTAAGCGTGGTGACAAAATTGTGTACCACACTCTGGAGAGCCCCGTCGAGTTTCATCTTGACGGTGAGGTTCTTTCACTTGACAAACTAAAGAGTCTCTTATCCCTGCGGGAGGTTAAGACTATAAAAGTGTTCACAACTGTGGACAACACTAATCTCCACACACAGCTTGTGGATATGTCTATGACATATGGACAGCAGTTTGGTCCAACATACTTGGATGGTGCTGATGTTACAAAAATTAAACCTCATGTAAATCATGAGGGTAAGACTTTCTTTGTACTACCTAGTGATGACACACTACGTAGTGAAGCTTTCGAGTACTACCATACTCTTGATGAGAGTTTTCTTGGTAGGTACATGTCTGCTTTAAACCACACAAAGAAATGGAAATTTCCTCAAGTTGGTGGTTTAACTTCAATTAAATGGGCTGATAACAATTGTTATTTGTCTAGTGTTTTATTAGCACTTCAACAGCTTGAAGTCAAATTCAATGCACCAGCACTTCAAGAGGCTTATTATAGAGCCCGTGCTGGTGATGCTGCTAACTTTTGTGCACTCATACTCGCTTACAGTAATAAAACTGTTGGCGAGCTTGGTGATGTCAGAGAAACTATGACCCATCTTCTACAGCATGCTAATTTGGAATCTGCAAAGCGAGTTCTTAATGTGGTGTGTAAACATTGTGGTCAGAAAACTACTACCTTAACGGGTGTAGAAGCTGTGATGTATATGGGTACTCTATCTTATGATAATCTTAAGACAGGTGTTTCCATTCCATGTGTGTGTGGTCGTGATGCTACACAATATCTAGTACAACAAGAGTCTTCTTTTGTTATGATGTCTGCACCACCTGCTGAGTATAAATTACAGCAAGGTACATTCTTATGTGCGAATGAGTACACTGGTAACTATCAGTGTGGTCATTACACTCATATAACTGCTAAGGAGACCCTCTATCGTATTGACGGAGCTCACCTTACAAAGATGTCAGAGTACAAAGGACCAGTGACTGATGTTTTCTACAAGGAAACATCTTACACTACAACCATCAAGCCTGTGTCGTATAAACTCGATGGAGTTACTTACACAGAGATTGAACCAAAATTGGATGGGTATTATAAAAAGGATAATGCTTACTATACAGAGCAGCCTATAGACCTTGTACCAACTCAACCATTACCAAATGCGAGTTTTGATAATTTCAAACTCACATGTTCTAACACAAAATTTGCTGATGATTTAAATCAAATGACAGGCTTCACAAAGCCAGCTTCACGAGAGCTATCTGTCACATTCTTCCCAGACTTGAATGGCGATGTAGTGGCTATTGACTATAGACACTATTCAGCGAGTTTCAAGAAAGGTGCTAAATTACTGCATAAGCCAATTGTTTGGCACATTAACCAGGCTACAACCAAGACAACGTTCAAACCAAACACTTGGTGTTTACGTTGTCTTTGGAGTACAAAGCCAGTAGATACTTCAAATTCATTTGAAGTTCTGGCAGTAGAAGACACACAAGGAATGGACAATCTTGCTTGTGAAAGTCAACAACCCACCTCTGAAGAAGTAGTGGAAAATCCTACCATACAGAAGGAAGTCATAGAGTGTGACGTGAAAACTACCGAAGTTGTAGGCAATGTCATACTTAAACCATCAGATGAAGGTGTTAAAGTAACACAAGAGTTAGGTCATGAGGATCTTATGGCTGCTTATGTGGAAAACACAAGCATTACCATTAAGAAACCTAATGAGCTTTCACTAGCCTTAGGTTTAAAAACAATTGCCACTCATGGTATTGCTGCAATTAATAGTGTTCCTTGGAGTAAAATTTTGGCTTATGTCAAACCATTCTTAGGACAAGCAGCAATTACAACATCAAATTGCGCTAAGAGATTAGCACAACGTGTGTTTAACAATTATATGCCTTATGTGTTTACATTATTGTTCCAATTGTGTACTTTTACTAAAAGTACCAATTCTAGAATTAGAGCTTCACTACCTACAACTATTGCTAAAAATAGTGTTAAGAGTGTTGCTAAATTATGTTTGGATGCCGGCATTAATTATGTGAAGTCACCCAAATTTTCTAAATTGTTCACAATCGCTATGTGGCTATTGTTGTTAAGTATTTGCTTAGGTTCTCTAATCTGTGTAACTGCTGCTTTTGGTGTACTCTTATCTAATTTTGGTGCTCCTTCTTATTGTAATGGCGTTAGAGAATTGTATCTTAATTCGTCTAACGTTACTACTATGGATTTCTGTGAAGGTTCTTTTCCTTGCAGCATTTGTTTAAGTGGATTAGACTCCCTTGATTCTTATCCAGCTCTTGAAACCATTCAGGTGACGATTTCATCGTACAAGCTAGACTTGACAATTTTAGGTCTGGCCGCTGAGTGGGTTTTGGCATATATGTTGTTCACAAAATTCTTTTATTTATTAGGTCTTTCAGCTATAATGCAGGTGTTCTTTGGCTATTTTGCTAGTCATTTCATCAGCAATTCTTGGCTCATGTGGTTTATCATTAGTATTGTACAAATGGCACCCGTTTCTGCAATGGTTAGGATGTACATCTTCTTTGCTTCTTTCTACTACATATGGAAGAGCTATGTTCATATCATGGATGGTTGCACCTCTTCGACTTGCATGATGTGCTATAAGCGCAATCGTGCCACACGCGTTGAGTGTACAACTATTGTTAATGGCATGAAGAGATCTTTCTATGTCTATGCAAATGGAGGCCGTGGCTTCTGCAAGACTCACAATTGGAATTGTCTCAATTGTGACACATTTTGCACTGGTAGTACATTCATTAGTGATGAAGTTGCTCGTGATTTGTCACTCCAGTTTAAAAGACCAATCAACCCTACTGACCAGTCATCGTATATTGTTGATAGTGTTGCTGTGAAAAATGGCGCGCTTCACCTCTACTTTGACAAGGCTGGTCAAAAGACCTATGAGAGACATCCGCTCTCCCATTTTGTCAATTTAGACAATTTGAGAGCTAACAACACTAAAGGTTCACTGCCTATTAATGTCATAGTTTTTGATGGCAAGTCCAAATGCGACGAGTCTGCTTCTAAGTCTGCTTCTGTGTACTACAGTCAGCTGATGTGCCAACCTATTCTGTTGCTTGACCAAGCTCTTGTATCAGACGTTGGAGATAGTACTGAAGTTTCCGTTAAGATGTTTGATGCTTATGTCGACACCTTTTCAGCAACTTTTAGTGTTCCTATGGAAAAACTTAAGGCACTTGTTGCTACAGCTCACAGCGAGTTAGCAAAGGGTGTAGCTTTAGATGGTGTCCTTTCTACATTCGTGTCAGCTGCCCGACAAGGTGTTGTTGATACCGATGTTGACACAAAGGATGTTATTGAATGTCTCAAACTTTCACATCACTCTGACTTAGAAGTGACAGGTGACAGTTGTAACAATTTCATGCTCACCTATAATAAGGTTGAAAACATGACGCCCAGAGATCTTGGCGCATGTATTGACTGTAATGCAAGGCATATCAATGCCCAAGTAGCAAAAAGTCACAATGTTTCACTCATCTGGAATGTAAAAGACTACATGTCTTTATCTGAACAGCTGCGTAAACAAATTCGTAGTGCTGCCAAGAAGAACAACATACCTTTTAGACTAACTTGTGCTACAACTAGACAGGTTGTCAATGTCATAACTACTAAAATCTCACTCAAGGGTGGTAAGATTGTTAGTACTTGTTTTAAACTTATGCTTAAGGCCACATTATTGTGCGTTCTTGCTGCATTGGTTTGTTATATCGTTATGCCAGTACATACATTGTCAATCCATGATGGTTACACAAATGAAATCATTGGTTACAAAGCCATTCAGGATGGTGTCACTCGTGACATCATTTCTACTGATGATTGTTTTGCAAATAAACATGCTGGTTTTGACGCATGGTTTAGCCAGCGTGGTGGTTCATACAAAAATGACAAAAGCTGCCCTGTAGTAGCTGCTATCATTACAAGAGAGATTGGTTTCATAGTGCCTGGCTTACCGGGTACTGTGCTGAGAGCAATCAATGGTGACTTCTTGCATTTTCTACCTCGTGTTTTTAGTGCTGTTGGCAACATTTGCTACACACCTTCCAAACTCATTGAGTATAGTGATTTTGCTACCTCTGCTTGCGTTCTTGCTGCTGAGTGTACAATTTTTAAGGATGCTATGGGCAAACCTGTGCCATATTGTTATGACACTAATTTGCTAGAGGGTTCTATTTCTTATAGTGAGCTTCGTCCAGACACTCGTTATGTGCTTATGGATGGTTCCATCATACAGTTTCCTAACACTTACCTGGAGGGTTCTGTTAGAGTAGTAACAACTTTTGATGCTGAGTACTGTAGACATGGTACATGCGAAAGGTCAGAAGTAGGTATTTGCCTATCTACCAGTGGTAGATGGGTTCTTAATAATGAGCATTACAGAGCTCTATCAGGAGTTTTCTGTGGTGTTGATGCGATGAATCTCATAGCTAACATCTTTACTCCTCTTGTGCAACCTGTGGGTGCTTTAGATGTGTCTGCTTCAGTAGTGGCTGGTGGTATTATTGCCATATTGGTGACTTGTGCTGCCTACTACTTTATGAAATTCAGACGTGTTTTTGGTGAGTACAACCATGTTGTTGCTGCTAATGCACTTTTGTTTTTGATGTCTTTCACTATACTCTGTCTGGTACCAGCTTACAGCTTTCTGCCGGGAGTCTACTCAGTCTTTTACTTGTACTTGACATTCTATTTCACCAATGATGTTTCATTCTTGGCTCACCTTCAATGGTTTGCCATGTTTTCTCCTATTGTGCCTTTTTGGATAACAGCAATCTATGTATTCTGTATTTCTCTGAAGCACTGCCATTGGTTCTTTAACAACTATCTTAGGAAAAGAGTCATGTTTAATGGAGTTACATTTAGTACCTTCGAGGAGGCTGCTTTGTGTACCTTTTTGCTCAACAAGGAAATGTACCTAAAATTGCGTAGCGAGACACTGTTGCCACTTACACAGTATAACAGGTATCTTGCTCTATATAACAAGTACAAGTATTTCAGTGGAGCCTTAGATACTACCAGCTATCGTGAAGCAGCTTGCTGCCACTTAGCAAAGGCTCTAAATGACTTTAGCAACTCAGGTGCTGATGTTCTCTACCAACCACCACAGACATCAATCACTTCTGCTGTTCTGCAGAGTGGTTTTAGGAAAATGGCATTCCCGTCAGGCAAAGTTGAAGGGTGCATGGTACAAGTAACCTGTGGAACTACAACTCTTAATGGATTGTGGTTGGATGACACAGTATACTGTCCAAGACATGTCATTTGCACAGCAGAAGACATGCTTAATCCTAACTATGAAGATCTGCTCATTCGCAAATCCAACCATAGCTTTCTTGTTCAGGCTGGCAATGTTCAACTTCGTGTTATTGGCCATTCTATGCAAAATTGTCTGCTTAGGCTTAAAGTTGATACTTCTAACCCTAAGACACCCAAGTATAAATTTGTCCGTATCCAACCTGGTCAAACATTTTCAGTTCTAGCATGCTACAATGGTTCACCATCTGGTGTTTATCAGTGTGCCATGAGACCTAATCATACCATTAAAGGTTCTTTCCTTAATGGATCATGTGGTAGTGTTGGTTTTAACATTGATTATGATTGCGTGTCTTTCTGCTATATGCATCATATGGAGCTTCCAACAGGAGTACACGCTGGTACTGACTTAGAAGGTAAATTCTATGGTCCATTTGTTGACAGACAAACTGCACAGGCTGCAGGTACAGACACAACCATAACATTAAATGTTTTGGCATGGCTGTATGCTGCTGTTATCAATGGTGATAGGTGGTTTCTTAATAGATTCACCACTACTTTGAATGACTTTAACCTTGTGGCAATGAAGTACAACTATGAACCTTTGACACAAGATCATGTTGACATATTGGGACCTCTTTCTGCTCAAACAGGAATTGCCGTCTTAGATATGTGTGCTGCTTTGAAAGAGCTGCTGCAGAATGGTATGAATGGTCGTACTATCCTTGGTAGCACTATTTTAGAAGATGAGTTTACACCATTTGATGTTGTTAGACAATGCTCTGGTGTTACCTTCCAAGGTAAGTTCAAGAAAATTGTTAAGGGCACTCATCATTGGATGCTTTTAACTTTCTTGACATCACTATTGATTCTTGTTCAAAGTACACAGTGGTCACTGTTTTTCTTTGTTTACGAGAATGCTTTCTTGCCATTTACTCTTGGTATTATGGCAATTGCTGCATGTGCTATGCTGCTTGTTAAGCATAAGCACGCATTCTTGTGCTTGTTTCTGTTACCTTCTCTTGCAACAGTTGCTTACTTTAATATGGTCTACATGCCTGCTAGCTGGGTGATGCGTATCATGACATGGCTTGAATTGGCTGACACTAGCTTGTCTGGTTATAGGCTTAAGGATTGTGTTATGTATGCTTCAGCTTTAGTTTTGCTTATTCTCATGACAGCTCGCACTGTTTATGATGATGCTGCTAGACGTGTTTGGACACTGATGAATGTCATTACACTTGTTTACAAAGTCTACTATGGTAATGCTTTAGATCAAGCTATTTCCATGTGGGCCTTAGTTATTTCTGTAACCTCTAACTATTCTGGTGTCGTTACGACTATCATGTTTTTAGCTAGAGCTATAGTGTTTGTGTGTGTTGAGTATTACCCATTGTTATTTATTACTGGCAACACCTTACAGTGTATCATGCTTGTTTATTGTTTCTTAGGCTATTGTTGCTGCTGCTACTTTGGCCTTTTCTGTTTACTCAACCGTTACTTCAGGCTTACTCTTGGTGTTTATGACTACTTGGTCTCTACACAAGAATTTAGGTATATGAACTCCCAGGGGCTTTTGCCTCCTAAGAGTAGTATTGATGCTTTCAAGCTTAACATTAAGTTGTTGGGTATTGGAGGTAAACCATGTATCAAGGTTGCTACTGTACAGTCTAAAATGTCTGACGTAAAGTGCACATCTGTGGTACTGCTCTCGGTTCTTCAACAACTTAGAGTAGAGTCATCTTCTAAATTGTGGGCACAATGTGTACAACTCCACAATGATATTCTTCTTGCAAAAGACACAACTGAAGCTTTCGAGAAGATGGTTTCTCTTTTGTCTGTTTTGCTATCCATGCAGGGTGCTGTAGACATTAATAGGTTGTGCGAGGAAATGCTCGATAACCGTGCTACTCTTCAGGCTATTGCTTCAGAATTTAGTTCTTTACCATCATATGCCGCTTATGCCACTGCCCAGGAGGCCTATGAGCAGGCTGTAGCTAATGGTGATTCTGAAGTCGTTCTCAAAAAGTTAAAGAAATCTTTGAATGTGGCTAAATCTGAGTTTGACCGTGATGCTGCCATGCAACGCAAGTTGGAAAAGATGGCAGATCAGGCTATGACCCAAATGTACAAACAGGCAAGATCTGAGGACAAGAGGGCAAAAGTAACTAGTGCTATGCAAACAATGCTCTTCACTATGCTTAGGAAGCTTGATAATGATGCACTTAACAACATTATCAACAATGCGCGTGATGGTTGTGTTCCACTCAACATCATACCATTGACTACAGCAGCCAAACTCATGGTTGTTGTCCCTGATTATGGTACCTACAAGAACACTTGTGATGGTAACACCTTTACATATGCATCTGCACTCTGGGAAATCCAGCAAGTTGTTGATGCGGATAGCAAGATTGTTCAACTTAGTGAAATTAACATGGACAATTCACCAAATTTGGCTTGGCCTCTTATTGTTACAGCTCTAAGAGCCAACTCAGCTGTTAAACTACAGAATAATGAACTGAGTCCAGTAGCACTACGACAGATGTCCTGTGCGGCTGGTACCACACAAACAGCTTGTACTGATGACAATGCACTTGCCTACTATAACAATTCGAAGGGAGGTAGGTTTGTGCTGGCATTACTATCAGACCACCAAGATCTCAAATGGGCTAGATTCCCTAAGAGTGATGGTACAGGTACAATTTACACAGAACTGGAACCACCTTGTAGGTTTGTTACAGACACACCAAAAGGGCCTAAAGTGAAATACTTGTACTTCATCAAAGGCTTAAACAACCTAAATAGAGGTATGGTGCTGGGCAGTTTAGCTGCTACAGTACGTCTTCAGGCTGGAAATGCTACAGAAGTACCTGCCAATTCAACTGTGCTTTCCTTCTGTGCTTTTGCAGTAGACCCTGCTAAAGCATATAAGGATTACCTAGCAAGTGGAGGACAACCAATCACCAACTGTGTGAAGATGTTGTGTACACACACTGGTACAGGACAGGCAATTACTGTAACACCAGAAGCTAACATGGACCAAGAGTCCTTTGGTGGTGCTTCATGTTGTCTGTATTGTAGATGCCACATTGACCATCCAAATCCTAAAGGATTCTGTGACTTGAAAGGTAAGTACGTCCAAATACCTACCACTTGTGCTAATGACCCAGTGGGTTTTACACTTAGAAACACAGTCTGTACCGTCTGCGGAATGTGGAAAGGTTATGGCTGTAGTTGTGACCAACTCCGCGAACCCTTGATGCAGTCTGCGGATGCATCAACGTTTTTAAACGGGTTTGCGGTGTAAGTGCAGCCCGTCTTACACCGTGCGGCACAGGCACTAGTACTGATGTCGTCTACAGGGCTTTTGATATTTACAACGAAAAAGTTGCTGGTTTTGCAAAGTTCCTAAAAACTAATTGCTGTCGCTTCCAGGAGAAGGATGAGGAAGGCAATTTATTAGACTCTTACTTTGTAGTTAAGAGGCATACTATGTCTAACTACCAACATGAAGAGACTATTTATAACTTGGTTAAAGATTGTCCAGCGGTTGCTGTCCATGACTTTTTCAAGTTTAGAGTAGATGGTGACATGGTACCACATATATCACGTCAGCGTCTAACTAAATACACAATGGCTGATTTAGTCTATGCTCTACGTCATTTTGATGAGGGTAATTGTGATACATTAAAAGAAATACTCGTCACATACAATTGCTGTGATGATGATTATTTCAATAAGAAGGATTGGTATGACTTCGTAGAGAATCCTGACATCTTACGCGTATATGCTAACTTAGGTGAGCGTGTACGCCAATCATTATTAAAGACTGTACAATTCTGCGATGCTATGCGTGATGCAGGCATTGTAGGCGTACTGACATTAGATAATCAGGATCTTAATGGGAACTGGTACGATTTCGGTGATTTCGTACAAGTAGCACCAGGCTGCGGAGTTCCTATTGTGGATTCATATTACTCATTGCTGATGCCCATCCTCACTTTGACTAGGGCATTGGCTGCTGAGTCCCATATGGATGCTGATCTCGCAAAACCACTTATTAAGTGGGATTTGCTGAAATATGATTTTACGGAAGAGAGACTTTGTCTCTTCGACCGTTATTTTAAATATTGGGACCAGACATACCATCCCAATTGTATTAACTGTTTGGATGATAGGTGTATCCTTCATTGTGCAAACTTTAATGTGTTATTTTCTACTGTGTTTCCACCTACAAGTTTTGGACCACTAGTAAGAAAAATATTTGTAGATGGTGTTCCTTTTGTTGTTTCAACTGGATACCATTTTCGTGAGTTAGGAGTCGTACATAATCAGGATGTAAACTTACATAGCTCGCGTCTCAGTTTCAAGGAACTTTTAGTGTATGCTGCTGATCCAGCTATGCATGCAGCTTCTGGCAATTTATTGCTAGATAAACGCACTACATGCTTTTCAGTAGCTGCACTAACAAACAATGTTGCTTTTCAAACTGTCAAACCCGGTAATTTTAATAAAGACTTTTATGACTTTGCTGTGTCTAAAGGTTTCTTTAAGGAAGGAAGTTCTGTTGAACTAAAACACTTCTTCTTTGCTCAGGATGGCAACGCTGCTATCAGTGATTATGACTATTATCGTTATAATCTGCCAACAATGTGTGATATCAGACAACTCCTATTCGTAGTTGAAGTTGTTGATAAATACTTTGATTGTTACGATGGTGGCTGTATTAATGCCAACCAAGTAATCGTTAACAATCTGGATAAATCAGCTGGTTTCCCATTTAATAAATGGGGTAAGGCTAGACTTTATTATGACTCAATGAGTTATGAGGATCAAGATGCACTTTTCGCGTATACTAAGCGTAATGTCATCCCTACTATAACTCAAATGAATCTTAAGTATGCCATTAGTGCAAAGAATAGAGCTCGCACCGTAGCTGGTGTCTCTATCTGTAGTACTATGACAAATAGACAGTTTCATCAGAAATTATTGAAGTCAATAGCCGCCACTAGAGGAGCTACTGTGGTAATTGGAACAAGCAAGTTTTACGGTGGCTGGCATAATATGTTAAAAACTGTTTACAGTGATGTAGAAACTCCACACCTTATGGGTTGGGATTATCCAAAATGTGACAGAGCCATGCCTAACATGCTTAGGATAATGGCCTCTCTTGTTCTTGCTCGCAAACATAACACTTGCTGTAACTTATCACACCGTTTCTACAGGTTAGCTAACGAGTGTGCGCAAGTATTAAGTGAGATGGTCATGTGTGGCGGCTCACTATATGTTAAACCAGGTGGAACATCATCCGGTGATGCTACAACTGCTTATGCTAATAGTGTCTTTAACATTTGTCAAGCTGTTACAGCCAATGTAAATGCACTTCTTTCAACTGATGGTAATAAGATAGCTGACAAGTATGTCCGCAATCTACAACACAGGCTCTATGAGTGTCTCTATAGAAATAGGGATGTTGATCATGAATTCGTGGATGAGTTTTACGCTTACCTGCGTAAACATTTCTCCATGATGATTCTTTCTGATGATGCCGTTGTGTGCTATAACAGTAACTATGCGGCTCAAGGTTTAGTAGCTAGCATTAAGAACTTTAAGGCAGTTCTTTATTATCAAAATAATGTGTTCATGTCTGAGGCAAAATGTTGGACTGAGACTGACCTTACTAAAGGACCTCACGAATTTTGCTCACAGCATACAATGCTAGTTAAACAAGGAGATGATTACGTGTACCTGCCTTACCCAGATCCATCAAGAATATTAGGCGCAGGCTGTTTTGTCGATGATATTGTCAAAACAGATGGTACACTTATGATTGAAAGGTTCGTGTCACTGGCTATTGATGCTTACCCACTTACAAAACATCCTAATCAGGAGTATGCTGATGTCTTTCACTTGTATTTACAATACATTAGAAAGTTACATGATGAGCTTACTGGCCACATGTTGGACATGTATTCCGTAATGCTAACTAATGATAACACCTCACGGTACTGGGAACCTGAGTTTTATGAGGCTATGTACACACCACATACAGTCTTGCAGGCTGTAGGTGCTTGTGTATTGTGCAATTCACAGACTTCACTTCGTTGCGGTGCCTGTATTAGGAGACCATTCCTATGTTGCAAGTGCTGCTATGACCATGTCATTTCAACATCACACAAATTAGTGTTGTCTGTTAATCCCTATGTTTGCAATGCCCCAGGTTGTGATGTCACTGATGTGACACAACTGTATCTAGGAGGTATGAGCTATTATTGCAAGTCACATAAGCCTCCCATTAGTTTTCCATTATGTGCTAATGGTCAGGTTTTTGGTTTATACAAAAACACATGTGTAGGCAGTGACAATGTCACTGACTTCAATGCGATAGCAACATGTGATTGGACTAATGCTGGCGATTACATACTTGCCAACACTTGTACTGAGAGACTCAAGCTTTTCGCAGCAGAAACGCTCAAAGCCACTGAGGAAACATTTAAGCTGTCATATGGTATTGCCACTGTACGCGAAGTACTCTCTGACAGAGAATTGCATCTTTCATGGGAGGTTGGAAAACCTAGACCACCATTGAACAGAAACTATGTCTTTACTGGTTACCGTGTAACTAAAAATAGTAAAGTACAGATTGGAGAGTACACCTTTGAAAAAGGTGACTATGGTGATGCTGTTGTGTACAGAGGTACTACGACATACAAGTTGAATGTTGGTGATTACTTTGTGTTGACATCTCACACTGTAATGCCACTTAGTGCACCTACTCTAGTGCCACAAGAGCACTATGTGAGAATTACTGGCTTGTACCCAACACTCAACATCTCAGATGAGTTTTCTAGCAATGTTGCAAATTATCAAAAGGTCGGCATGCAAAAGTACTCTACACTCCAAGGACCACCTGGTACTGGTAAGAGTCATTTTGCCATCGGACTTGCTCTCTATTACCCATCTGCTCGCATAGTGTATACGGCATGCTCTCATGCAGCTGTTGATGCCCTATGTGAAAAGGCATTAAAATATTTGCCCATAGATAAATGTAGTAGAATCATACCTGCGCGTGCGCGCGTAGAGTGTTTTGATAAATTCAAAGTGAATTCAACACTAGAACAGTATGTTTTCTGCACTGTAAATGCATTGCCAGAAACAACTGCTGACATTGTAGTCTTTGATGAAATCTCTATGGCTACTAATTATGACTTGAGTGTTGTCAATGCTAGACTTCGTGCAAAACACTACGTCTATATTGGCGATCCTGCTCAATTACCAGCCCCCCGCACATTGCTGACTAAAGGCACACTAGAACCAGAATATTTTAATTCAGTGTGCAGACTTATGAAAACAATAGGTCCAGACATGTTCCTTGGAACTTGTCGCCGTTGTCCTGCTGAAATTGTTGACACTGTGAGTGCTTTAGTTTATGACAATAAGCTAAAAGCACACAAGGATAAGTCAGCTCAATGCTTCAAAATGTTCTACAAAGGTGTTATTACACATGATGTTTCATCTGCAATCAACAGACCTCAAATAGGCGTTGTAAGAGAATTTCTTACACGCAATCCTGCTTGGAGAAAAGCTGTTTTTATCTCACCTTATAATTCACAGAACGCTGTAGCTTCAAAAATCTTAGGATTGCCTACGCAGACTGTTGATTCATCACAGGGTTCTGAATATGACTATGTCATATTCACACAAACTACTGAAACAGCACACTCTTGTAATGTCAACCGCTTCAATGTGGCTATCACAAGGGCAAAAATTGGCATTTTGTGCATAATGTCTGATAGAGATCTTTATGACAAACTGCAATTTACAAGTCTAGAAATACCACGTCGCAATGTGGCTACATTACAAGCAGAAAATGTAACTGGACTTTTTAAGGACTGTAGTAAGATCATTACTGGTCTTCATCCTACACAGGCACCTACACACCTCAGCGTTGATATAAAGTTCAAGACTGAAGGATTATGTGTTGACATACCAGGCATACCAAAGGACATGACCTACCGTAGACTCATCTCTATGATGGGTTTCAAAATGAATTACCAAGTCAATGGTTACCCTAATATGTTTATCACCCGCGAAGAAGCTATTCGTCACGTTCGTGCGTGGATTGGCTTTGATGTAGAGGGCTGTCATGCAACTAGAGATGCTGTGGGTACTAACCTACCTCTCCAGCTAGGATTTTCTACAGGTGTTAACTTAGTAGCTGTACCGACTGGTTATGTTGACACTGAAAATAACACAGAATTCACCAGAGTTAATGCAAAACCTCCACCAGGTGACCAGTTTAAACATCTTATACCACTCATGTATAAAGGCTTGCCCTGGAATGTAGTGCGTATTAAGATAGTACAAATGCTCAGTGATACACTGAAAGGATTGTCAGACAGAGTCGTGTTCGTCCTTTGGGCGCATGGCTTTGAGCTTACATCAATGAAGTACTTTGTCAAGATTGGACCTGAAAGAACGTGTTGTCTGTGTGACAAACGTGCAACTTGCTTTTCTACTTCATCAGATACTTATGCCTGCTGGAATCATTCTGTGGGTTTTGACTATGTCTATAACCCATTTATGATTGATGTTCAGCAGTGGGGCTTTACGGGTAACCTTCAGAGTAACCATGACCAACATTGCCAGGTACATGGAAATGCACATGTGGCTAGTTGTGATGCTATCATGACTAGATGTTTAGCAGTCCATGAGTGCTTTGTTAAGCGCGTTGATTGGTCTGTTGAATACCCTATTATAGGAGATGAACTGAGGGTTAATTCTGCTTGCAGAAAAGTACAACACATGGTTGTGAAGTCTGCATTGCTTGCTGATAAGTTTCCAGTTCTTCATGACATTGGAAATCCAAAGGCTATCAAGTGTGTGCCTCAGGCTGAAGTAGAATGGAAGTTCTACGATGCTCAGCCATGTAGTGACAAAGCTTACAAAATAGAGGAACTCTTCTATTCTTATGCTACACATCACGATAAATTCACTGATGGTGTTTGTTTGTTTTGGAATTGTAACGTTGATCGTTACCCAGCCAATGCAATTGTGTGTAGGTTTGACACAAGAGTCTTGTCAAACTTGAACTTACCAGGCTGTGATGGTGGTAGTTTGTATGTGAATAAGCATGCATTCCACACTCCAGCTTTCGATAAAAGTGCATTTACTAATTTAAAGCAATTGCCTTTCTTTTACTATTCTGATAGTCCTTGTGAGTCTCATGGCAAACAAGTAGTGTCGGATATTGATTATGTTCCACTCAAATCTGCTACGTGTATTACACGATGCAATTTAGGTGGTGCTGTTTGCAGACACCATGCAAATGAGTACCGACAGTACTTGGATGCATATAATATGATGATTTCTGCTGGATTTAGCCTATGGATTTACAAACAATTTGATACTTATAACCTGTGGAATACATTTACCAGGTTACAGAGTTTAGAAAATGTGGCTTATAATGTTGTTAATAAAGGACACTTTGATGGACACGCCGGCGAAGCACCTGTTTCCATCATTAATAATGCTGTTTACACAAAGGTAGATGGTATTGATGTGGAGATCTTTGAAAATAAGACAACACTTCCTGTTAATGTTGCATTTGAGCTTTGGGCTAAGCGTAACATTAAACCAGTGCCAGAGATTAAGATACTCAATAATTTGGGTGTTGATATCGCTGCTAATACTGTAATCTGGGACTACAAAAGAGAAGCCCCAGCACATGTATCTACAATAGGTGTCTGCACAATGACTGACATTGCCAAGAAACCTACTGAGAGTGCTTGTTCTTCACTTACTGTCTTGTTTGATGGTAGAGTGGAAGGACAGGTAGACCTTTTTAGAAACGCCCGTAATGGTGTTTTAATAACAGAAGGTTCAGTCAAAGGTCTAACACCTTCAAAGGGACCAGCACAAGCTAGCGTCAATGGAGTCACATTAATTGGAGAATCAGTAAAAACACAGTTTAACTACTTTAAGAAAGTAGACGGCATTATTCAACAGTTGCCTGAAACCTACTTTACTCAGAGCAGAGACTTAGAGGATTTTAAGCCCAGATCACAAATGGAAACTGACTTTCTCGAGCTCGCTATGGATGAATTCATACAGCGATATAAGCTCGAGGGCTATGCCTTCGAACACATCGTTTATGGAGATTTCAGTCATGGACAACTTGGCGGTCTTCATTTAATGATAGGCTTAGCCAAGCGCTCACAAGATTCACCACTTAAATTAGAGGATTTTATCCCTATGGACAGCACAGTGAAAAATTACTTCATAACAGATGCGCAAACAGGTTCATCAAAATGTGTGTGTTCTGTGATTGATCTTTTACTTGATGACTTTGTCGAGATAATAAAGTCACAAGATTTGTCAGTGATTTCAAAAGTGGTCAAGGTTACAATTGACTATGCTGAAATTTCATTCATGCTTTGGTGTAAGGATGGACATGTTGAAACCTTCTACCCAAAACTACAAGCAAGTCAAGCGTGGCAACCAGGTGTTGCGATGCCTAACTTGTACAAGATGCAAAGAATGCTTCTTGAAAAGTGTGACCTTCAGAATTATGGTGAAAATGCTGTTATACCAAAAGGAATAATGATGAATGTCGCAAAGTATACTCAACTGTGTCAATACTTAAATACACTTACTTTAGCTGTACCCTACAACATGAGAGTTATTCACTTTGGTGCTGGCTCTGATAAAGGAGTTGCACCAGGTACAGCTGTGCTCAGACAATGGTTGCCAACTGGCACACTACTTGTCGATTCAGATCTTAATGACTTCGTCTCCGACGCAGATTCTACTTTAATTGGAGACTGTGCAACAGTACATACGGCTAATAAATGGGACCTTATTATTAGCGATATGTATGACCCTAGGACCAAACATGTGACAAAAGAGAATGACTCTAAAGAAGGGTTTTTCACTTATCTGTGTGGATTTATAAAGCAAAAACTAGCCCTGGGTGGTTCTATAGCTGTAAAGATAACAGAGCATTCTTGGAATGCTGACCTTTACAAGCTTATGGGCCATTTCTCATGGTGGACAGCTTTTGTTACAAATGTAAATGCATCATCATCGGAAGCATTTTTAATTGGGGCTAACTATCTTGGCAAGCCGAAGGAACAAATTGATGGCTATACCATGCATGCTAACTACATTTTCTGGAGGAACACAAATCCTATCCAGTTGTCTTCCTATTCACTCTTTGACATGAGCAAATTTCCTCTTAAATTAAGAGGAACTGCTGTAATGTCTCTTAAGGAGAATCAAATCAATGATATGATTTATTCTCTTCTGGAAAAAGGTAGGCTTATCATTAGAGAAAACAACAGAGTTGTGGTTTCAAGTGATATTCTTGTTAACAACTAA
Protein
MESLVLGVNEKTHVQLSLPVLQVRDVLVRGFGDSVEEALSEAREHLKNGTCGLVELEKGVLPQLEQPYVFIKRSDALSTNHGHKVVELVAEMDGIQYGRSGITLGVLVPHVGETPIAYRNVLLRKNGNKGAGGHSYGIDLKSYDLGDELGTDPIEDYEQNWNTKHGSGALRELTRELNGGAVTRYVDNNFCGPDGYPLDCIKDFLARAGKSMCTLSEQLDYIESKRGVYCCRDHEHEIAWFTERSDKSYEHQTPFEIKSAKKFDTFKGECPKFVFPLNSKVKVIQPRVEKKKTEGFMGRIRSVYPVASPQECNNMHLSTLMKCNHCDEVSWQTCDFLKATCEHCGTENLVIEGPTTCGYLPTNAVVKMPCPACQDPEIGPEHSVADYHNHSNIETRLRKGGRTRCFGGCVFAYVGCYNKRAYWVPRASADIGSGHTGITGDNVETLNEDLLEILSRERVNINIVGDFHLNEEVAIILASFSASTSAFIDTIKSLDYKSFKTIVESCGNYKVTKGKPVKGAWNIGQQRSVLTPLCGFPSQAAGVIRSIFARTLDAANHSIPDLQRAAVTILDGISEQSLRLVDAMVYTSDLLTNSVIIMAYVTGGLVQQTSQWLSNLLGTTVEKLRPIFEWIEAKLSAGVEFLKDAWEILKFLITGVFDIVKGQIQVASDNIKDCVKCFIDVVNKALEMCIDQVTIAGAKLRSLNLGEVFIAQSKGLYRQCIRGKEQLQLLMPLKAPKEVTFLEGDSHDTVLTSEEVVLKNGELEALETPVDSFTNGAIVGTPVCVNGLMLLEIKDKEQYCALSPGLLATNNVFRLKGGAPIKGVTFGEDTVWEVQGYKNVRITFELDERVDKVLNEKCSVYTVESGTEVTEFACVVAEAVVKTLQPVSDLLTNMGIDLDEWSVATFYLFDDAGEENFSSRMYCSFYPPDEEEEDDAECEEEEIDETCEHEYGTEDDYQGLPLEFGASAETVRVEEEEEEDWLDDTTEQSEIEPEPEPTPEEPVNQFTGYLKLTDNVAIKCVDIVKEAQSANPMVIVNAANIHLKHGGGVAGALNKATNGAMQKESDDYIKLNGPLTVGGSCLLSGHNLAKKCLHVVGPNLNAGEDIQLLKAAYENFNSQDILLAPLLSAGIFGAKPLQSLQVCVQTVRTQVYIAVNDKALYEQVVMDYLDNLKPRVEAPKQEEPPNTEDSKTEEKSVVQKPVDVKPKIKACIDEVTTTLEETKFLTNKLLLFADINGKLYHDSQNMLRGEDMSFLEKDAPYMVGDVITSGDITCVVIPSKKAGGTTEMLSRALKKVPVDEYITTYPGQGCAGYTLEEAKTALKKCKSAFYVLPSEAPNAKEEILGTVSWNLREMLAHAEETRKLMPICMDVRAIMATIQRKYKGIKIQEGIVDYGVRFFFYTSKEPVASIITKLNSLNEPLVTMPIGYVTHGFNLEEAARCMRSLKAPAVVSVSSPDAVTTYNGYLTSSSKTSEEHFVETVSLAGSYRDWSYSGQRTELGVEFLKRGDKIVYHTLESPVEFHLDGEVLSLDKLKSLLSLREVKTIKVFTTVDNTNLHTQLVDMSMTYGQQFGPTYLDGADVTKIKPHVNHEGKTFFVLPSDDTLRSEAFEYYHTLDESFLGRYMSALNHTKKWKFPQVGGLTSIKWADNNCYLSSVLLALQQLEVKFNAPALQEAYYRARAGDAANFCALILAYSNKTVGELGDVRETMTHLLQHANLESAKRVLNVVCKHCGQKTTTLTGVEAVMYMGTLSYDNLKTGVSIPCVCGRDATQYLVQQESSFVMMSAPPAEYKLQQGTFLCANEYTGNYQCGHYTHITAKETLYRIDGAHLTKMSEYKGPVTDVFYKETSYTTTIKPVSYKLDGVTYTEIEPKLDGYYKKDNAYYTEQPIDLVPTQPLPNASFDNFKLTCSNTKFADDLNQMTGFTKPASRELSVTFFPDLNGDVVAIDYRHYSASFKKGAKLLHKPIVWHINQATTKTTFKPNTWCLRCLWSTKPVDTSNSFEVLAVEDTQGMDNLACESQQPTSEEVVENPTIQKEVIECDVKTTEVVGNVILKPSDEGVKVTQELGHEDLMAAYVENTSITIKKPNELSLALGLKTIATHGIAAINSVPWSKILAYVKPFLGQAAITTSNCAKRLAQRVFNNYMPYVFTLLFQLCTFTKSTNSRIRASLPTTIAKNSVKSVAKLCLDAGINYVKSPKFSKLFTIAMWLLLLSICLGSLICVTAAFGVLLSNFGAPSYCNGVRELYLNSSNVTTMDFCEGSFPCSICLSGLDSLDSYPALETIQVTISSYKLDLTILGLAAEWVLAYMLFTKFFYLLGLSAIMQVFFGYFASHFISNSWLMWFIISIVQMAPVSAMVRMYIFFASFYYIWKSYVHIMDGCTSSTCMMCYKRNRATRVECTTIVNGMKRSFYVYANGGRGFCKTHNWNCLNCDTFCTGSTFISDEVARDLSLQFKRPINPTDQSSYIVDSVAVKNGALHLYFDKAGQKTYERHPLSHFVNLDNLRANNTKGSLPINVIVFDGKSKCDESASKSASVYYSQLMCQPILLLDQALVSDVGDSTEVSVKMFDAYVDTFSATFSVPMEKLKALVATAHSELAKGVALDGVLSTFVSAARQGVVDTDVDTKDVIECLKLSHHSDLEVTGDSCNNFMLTYNKVENMTPRDLGACIDCNARHINAQVAKSHNVSLIWNVKDYMSLSEQLRKQIRSAAKKNNIPFRLTCATTRQVVNVITTKISLKGGKIVSTCFKLMLKATLLCVLAALVCYIVMPVHTLSIHDGYTNEIIGYKAIQDGVTRDIISTDDCFANKHAGFDAWFSQRGGSYKNDKSCPVVAAIITREIGFIVPGLPGTVLRAINGDFLHFLPRVFSAVGNICYTPSKLIEYSDFATSACVLAAECTIFKDAMGKPVPYCYDTNLLEGSISYSELRPDTRYVLMDGSIIQFPNTYLEGSVRVVTTFDAEYCRHGTCERSEVGICLSTSGRWVLNNEHYRALSGVFCGVDAMNLIANIFTPLVQPVGALDVSASVVAGGIIAILVTCAAYYFMKFRRVFGEYNHVVAANALLFLMSFTILCLVPAYSFLPGVYSVFYLYLTFYFTNDVSFLAHLQWFAMFSPIVPFWITAIYVFCISLKHCHWFFNNYLRKRVMFNGVTFSTFEEAALCTFLLNKEMYLKLRSETLLPLTQYNRYLALYNKYKYFSGALDTTSYREAACCHLAKALNDFSNSGADVLYQPPQTSITSAVLQSGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDTVYCPRHVICTAEDMLNPNYEDLLIRKSNHSFLVQAGNVQLRVIGHSMQNCLLRLKVDTSNPKTPKYKFVRIQPGQTFSVLACYNGSPSGVYQCAMRPNHTIKGSFLNGSCGSVGFNIDYDCVSFCYMHHMELPTGVHAGTDLEGKFYGPFVDRQTAQAAGTDTTITLNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKYNYEPLTQDHVDILGPLSAQTGIAVLDMCAALKELLQNGMNGRTILGSTILEDEFTPFDVVRQCSGVTFQGKFKKIVKGTHHWMLLTFLTSLLILVQSTQWSLFFFVYENAFLPFTLGIMAIAACAMLLVKHKHAFLCLFLLPSLATVAYFNMVYMPASWVMRIMTWLELADTSLSGYRLKDCVMYASALVLLILMTARTVYDDAARRVWTLMNVITLVYKVYYGNALDQAISMWALVISVTSNYSGVVTTIMFLARAIVFVCVEYYPLLFITGNTLQCIMLVYCFLGYCCCCYFGLFCLLNRYFRLTLGVYDYLVSTQEFRYMNSQGLLPPKSSIDAFKLNIKLLGIGGKPCIKVATVQSKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILLAKDTTEAFEKMVSLLSVLLSMQGAVDINRLCEEMLDNRATLQAIASEFSSLPSYAAYATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLEKMADQAMTQMYKQARSEDKRAKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNIIPLTTAAKLMVVVPDYGTYKNTCDGNTFTYASALWEIQQVVDADSKIVQLSEINMDNSPNLAWPLIVTALRANSAVKLQNNELSPVALRQMSCAAGTTQTACTDDNALAYYNNSKGGRFVLALLSDHQDLKWARFPKSDGTGTIYTELEPPCRFVTDTPKGPKVKYLYFIKGLNNLNRGMVLGSLAATVRLQAGNATEVPANSTVLSFCAFAVDPAKAYKDYLASGGQPITNCVKMLCTHTGTGQAITVTPEANMDQESFGGASCCLYCRCHIDHPNPKGFCDLKGKYVQIPTTCANDPVGFTLRNTVCTVCGMWKGYGCSCDQLREPLMQSADASTFLNRVCGVSAARLTPCGTGTSTDVVYRAFDIYNEKVAGFAKFLKTNCCRFQEKDEEGNLLDSYFVVKRHTMSNYQHEETIYNLVKDCPAVAVHDFFKFRVDGDMVPHISRQRLTKYTMADLVYALRHFDEGNCDTLKEILVTYNCCDDDYFNKKDWYDFVENPDILRVYANLGERVRQSLLKTVQFCDAMRDAGIVGVLTLDNQDLNGNWYDFGDFVQVAPGCGVPIVDSYYSLLMPILTLTRALAAESHMDADLAKPLIKWDLLKYDFTEERLCLFDRYFKYWDQTYHPNCINCLDDRCILHCANFNVLFSTVFPPTSFGPLVRKIFVDGVPFVVSTGYHFRELGVVHNQDVNLHSSRLSFKELLVYAADPAMHAASGNLLLDKRTTCFSVAALTNNVAFQTVKPGNFNKDFYDFAVSKGFFKEGSSVELKHFFFAQDGNAAISDYDYYRYNLPTMCDIRQLLFVVEVVDKYFDCYDGGCINANQVIVNNLDKSAGFPFNKWGKARLYYDSMSYEDQDALFAYTKRNVIPTITQMNLKYAISAKNRARTVAGVSICSTMTNRQFHQKLLKSIAATRGATVVIGTSKFYGGWHNMLKTVYSDVETPHLMGWDYPKCDRAMPNMLRIMASLVLARKHNTCCNLSHRFYRLANECAQVLSEMVMCGGSLYVKPGGTSSGDATTAYANSVFNICQAVTANVNALLSTDGNKIADKYVRNLQHRLYECLYRNRDVDHEFVDEFYAYLRKHFSMMILSDDAVVCYNSNYAAQGLVASIKNFKAVLYYQNNVFMSEAKCWTETDLTKGPHEFCSQHTMLVKQGDDYVYLPYPDPSRILGAGCFVDDIVKTDGTLMIERFVSLAIDAYPLTKHPNQEYADVFHLYLQYIRKLHDELTGHMLDMYSVMLTNDNTSRYWEPEFYEAMYTPHTVLQAVGACVLCNSQTSLRCGACIRRPFLCCKCCYDHVISTSHKLVLSVNPYVCNAPGCDVTDVTQLYLGGMSYYCKSHKPPISFPLCANGQVFGLYKNTCVGSDNVTDFNAIATCDWTNAGDYILANTCTERLKLFAAETLKATEETFKLSYGIATVREVLSDRELHLSWEVGKPRPPLNRNYVFTGYRVTKNSKVQIGEYTFEKGDYGDAVVYRGTTTYKLNVGDYFVLTSHTVMPLSAPTLVPQEHYVRITGLYPTLNISDEFSSNVANYQKVGMQKYSTLQGPPGTGKSHFAIGLALYYPSARIVYTACSHAAVDALCEKALKYLPIDKCSRIIPARARVECFDKFKVNSTLEQYVFCTVNALPETTADIVVFDEISMATNYDLSVVNARLRAKHYVYIGDPAQLPAPRTLLTKGTLEPEYFNSVCRLMKTIGPDMFLGTCRRCPAEIVDTVSALVYDNKLKAHKDKSAQCFKMFYKGVITHDVSSAINRPQIGVVREFLTRNPAWRKAVFISPYNSQNAVASKILGLPTQTVDSSQGSEYDYVIFTQTTETAHSCNVNRFNVAITRAKIGILCIMSDRDLYDKLQFTSLEIPRRNVATLQAENVTGLFKDCSKIITGLHPTQAPTHLSVDIKFKTEGLCVDIPGIPKDMTYRRLISMMGFKMNYQVNGYPNMFITREEAIRHVRAWIGFDVEGCHATRDAVGTNLPLQLGFSTGVNLVAVPTGYVDTENNTEFTRVNAKPPPGDQFKHLIPLMYKGLPWNVVRIKIVQMLSDTLKGLSDRVVFVLWAHGFELTSMKYFVKIGPERTCCLCDKRATCFSTSSDTYACWNHSVGFDYVYNPFMIDVQQWGFTGNLQSNHDQHCQVHGNAHVASCDAIMTRCLAVHECFVKRVDWSVEYPIIGDELRVNSACRKVQHMVVKSALLADKFPVLHDIGNPKAIKCVPQAEVEWKFYDAQPCSDKAYKIEELFYSYATHHDKFTDGVCLFWNCNVDRYPANAIVCRFDTRVLSNLNLPGCDGGSLYVNKHAFHTPAFDKSAFTNLKQLPFFYYSDSPCESHGKQVVSDIDYVPLKSATCITRCNLGGAVCRHHANEYRQYLDAYNMMISAGFSLWIYKQFDTYNLWNTFTRLQSLENVAYNVVNKGHFDGHAGEAPVSIINNAVYTKVDGIDVEIFENKTTLPVNVAFELWAKRNIKPVPEIKILNNLGVDIAANTVIWDYKREAPAHVSTIGVCTMTDIAKKPTESACSSLTVLFDGRVEGQVDLFRNARNGVLITEGSVKGLTPSKGPAQASVNGVTLIGESVKTQFNYFKKVDGIIQQLPETYFTQSRDLEDFKPRSQMETDFLELAMDEFIQRYKLEGYAFEHIVYGDFSHGQLGGLHLMIGLAKRSQDSPLKLEDFIPMDSTVKNYFITDAQTGSSKCVCSVIDLLLDDFVEIIKSQDLSVISKVVKVTIDYAEISFMLWCKDGHVETFYPKLQASQAWQPGVAMPNLYKMQRMLLEKCDLQNYGENAVIPKGIMMNVAKYTQLCQYLNTLTLAVPYNMRVIHFGAGSDKGVAPGTAVLRQWLPTGTLLVDSDLNDFVSDADSTLIGDCATVHTANKWDLIISDMYDPRTKHVTKENDSKEGFFTYLCGFIKQKLALGGSIAVKITEHSWNADLYKLMGHFSWWTAFVTNVNASSSEAFLIGANYLGKPKEQIDGYTMHANYIFWRNTNPIQLSSYSLFDMSKFPLKLRGTAVMSLKENQINDMIYSLLEKGRLIIRENNRVVVSSDILVNN
Summary
Function
Inhibits host translation by interacting with the 40S ribosomal subunit. The nsp1-40S ribosome complex further induces an endonucleolytic cleavage near the 5'UTR of host mRNAs, targeting them for degradation. Viral mRNAs are not susceptible to nsp1-mediated endonucleolytic RNA cleavage thanks to the presence of a 5'-end leader sequence and are therefore protected from degradation. By suppressing host gene expression, nsp1 facilitates efficient viral gene expression in infected cells and evasion from host immune response.
May play a role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2. Indeed, these two proteins play a role in maintaining the functional integrity of the mitochondria and protecting cells from various stresses.
Responsible for the cleavages located at the N-terminus of the replicase polyprotein. In addition, PL-PRO possesses a deubiquitinating/deISGylating activity and processes both 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains from cellular substrates. Participates together with nsp4 in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication. Antagonizes innate immune induction of type I interferon by blocking the phosphorylation, dimerization and subsequent nuclear translocation of host IRF3. Prevents also host NF-kappa-B signaling.
Participates in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication.
Proteinase 3CL-PRO: Ccleaves the C-terminus of replicase polyprotein at 11 sites. Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. Also able to bind an ADP-ribose-1''-phosphate (ADRP).
Plays a role in the initial induction of autophagosomes from host reticulum endoplasmic. Later, limits the expansion of these phagosomes that are no longer able to deliver viral components to lysosomes.
Forms a hexadecamer with nsp8 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
Forms a hexadecamer with nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
May participate in viral replication by acting as a ssRNA-binding protein.
Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease and nsp16 2'-O-methyltransferase activities. Therefore plays an essential role in viral mRNAs cap methylation.
Responsible for replication and transcription of the viral RNA genome.
Multi-functional protein with a zinc-binding domain in N-terminus displaying RNA and DNA duplex-unwinding activities with 5' to 3' polarity. Activity of helicase is dependent on magnesium.
Enzyme possessing two different activities: an exoribonuclease activity acting on both ssRNA and dsRNA in a 3' to 5' direction and a N7-guanine methyltransferase activity.
Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs. N7-methyl guanosine cap is a prerequisite for binding of nsp16. Therefore plays an essential role in viral mRNAs cap methylation which is essential to evade immune system.
May play a role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2. Indeed, these two proteins play a role in maintaining the functional integrity of the mitochondria and protecting cells from various stresses.
Responsible for the cleavages located at the N-terminus of the replicase polyprotein. In addition, PL-PRO possesses a deubiquitinating/deISGylating activity and processes both 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains from cellular substrates. Participates together with nsp4 in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication. Antagonizes innate immune induction of type I interferon by blocking the phosphorylation, dimerization and subsequent nuclear translocation of host IRF3. Prevents also host NF-kappa-B signaling.
Participates in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication.
Proteinase 3CL-PRO: Ccleaves the C-terminus of replicase polyprotein at 11 sites. Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. Also able to bind an ADP-ribose-1''-phosphate (ADRP).
Plays a role in the initial induction of autophagosomes from host reticulum endoplasmic. Later, limits the expansion of these phagosomes that are no longer able to deliver viral components to lysosomes.
Forms a hexadecamer with nsp8 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
Forms a hexadecamer with nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
May participate in viral replication by acting as a ssRNA-binding protein.
Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease and nsp16 2'-O-methyltransferase activities. Therefore plays an essential role in viral mRNAs cap methylation.
Responsible for replication and transcription of the viral RNA genome.
Multi-functional protein with a zinc-binding domain in N-terminus displaying RNA and DNA duplex-unwinding activities with 5' to 3' polarity. Activity of helicase is dependent on magnesium.
Enzyme possessing two different activities: an exoribonuclease activity acting on both ssRNA and dsRNA in a 3' to 5' direction and a N7-guanine methyltransferase activity.
Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs. N7-methyl guanosine cap is a prerequisite for binding of nsp16. Therefore plays an essential role in viral mRNAs cap methylation which is essential to evade immune system.
Catalytic Activity
a ribonucleoside 5'-triphosphate + RNA(n) = diphosphate + RNA(n+1)
ATP + H2O = ADP + H(+) + phosphate
TSAVLQ-|-SGFRK-NH(2) and SGVTFQ-|-GKFKK the two peptides corresponding to the two self-cleavage sites of the SARS 3C-like proteinase are the two most reactive peptide substrates. The enzyme exhibits a strong preference for substrates containing Gln at P1 position and Leu at P2 position.
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
ATP + H2O = ADP + H(+) + phosphate
TSAVLQ-|-SGFRK-NH(2) and SGVTFQ-|-GKFKK the two peptides corresponding to the two self-cleavage sites of the SARS 3C-like proteinase are the two most reactive peptide substrates. The enzyme exhibits a strong preference for substrates containing Gln at P1 position and Leu at P2 position.
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Biophysicochemical Properties
1.15 mM for peptide TSAVLQ/SGFRK-NH(2)
0.58 mM for peptide SGVTFQ/GKFKK
1.44 mM for peptide ATVRLQ/AGNAT
0.58 mM for peptide SGVTFQ/GKFKK
1.44 mM for peptide ATVRLQ/AGNAT
Subunit
Nsp2 interacts with host PHB and PHB2. 3CL-PRO exists as monomer and homodimer. Nsp4 interacts with PL-PRO and nsp6. Only the homodimer shows catalytic activity. Eight copies of nsp7 and eight copies of nsp8 assemble to form a heterohexadecamer dsRNA-encircling ring structure. Nsp9 is a dimer. Nsp10 forms a dodecamer and interacts with nsp14 and nsp16; these interactions enhance nsp14 and nsp16 enzymatic activities. Nsp14 interacts (via N-terminus) with DDX1.
Miscellaneous
Produced by -1 ribosomal frameshifting at the 1a-1b genes boundary.
Similarity
Belongs to the coronaviruses polyprotein 1ab family.
Keywords
3D-structure
Activation of host autophagy by virus
ATP-binding
Decay of host mRNAs by virus
Endonuclease
Eukaryotic host gene expression shutoff by virus
Eukaryotic host translation shutoff by virus
Exonuclease
Helicase
Host cytoplasm
Host gene expression shutoff by virus
Host membrane
Host mRNA suppression by virus
Host-virus interaction
Hydrolase
Inhibition of host innate immune response by virus
Inhibition of host interferon signaling pathway by virus
Inhibition of host ISG15 by virus
Inhibition of host NF-kappa-B by virus
Membrane
Metal-binding
Methyltransferase
Modulation of host ubiquitin pathway by viral deubiquitinase
Modulation of host ubiquitin pathway by virus
Nuclease
Nucleotide-binding
Nucleotidyltransferase
Protease
Reference proteome
Repeat
Ribosomal frameshifting
RNA-binding
RNA-directed RNA polymerase
Thiol protease
Transferase
Transmembrane
Transmembrane helix
Ubl conjugation pathway
Viral immunoevasion
Viral RNA replication
Zinc
Zinc-finger
Feature
chain Host translation inhibitor nsp1
Uniprot
Pubmed
12730500
12730501
12853594
12958366
12876307
12781537
+ More
14527350 14645828 12917423 12917450 14561748 15331731 15564471 16549795 16882730 17024178 17692280 17855519 18417574 18827877 19369340 19640993 19153232 20573827 20421945 22174690 21345958 22039154 23035226 22615777 22791111 22635272 23943763 24991833 12746549 12925794 15007178 14962394 16228002 16271890 16581910 16873246 16873247 17202208
14527350 14645828 12917423 12917450 14561748 15331731 15564471 16549795 16882730 17024178 17692280 17855519 18417574 18827877 19369340 19640993 19153232 20573827 20421945 22174690 21345958 22039154 23035226 22615777 22791111 22635272 23943763 24991833 12746549 12925794 15007178 14962394 16228002 16271890 16581910 16873246 16873247 17202208
EMBL
AY278741
AY274119
AY278554
AY282752
AY304495
AY304486
+ More
AY304488 AY278491 AY283794 AY283795 AY283796 AY283797 AY283798 AY286320 AY278488 AY278489 AY278490 AY279354 AY291451 AY310120 AY291315 AY323977 AY321118 AY338174 AY338175 AY348314 AP006557 AP006558 AP006559 AP006560 AP006561 AY427439 AY322205 AY322206 AY322207 AY463059 AY269391 AY268049
AY304488 AY278491 AY283794 AY283795 AY283796 AY283797 AY283798 AY286320 AY278488 AY278489 AY278490 AY279354 AY291451 AY310120 AY291315 AY323977 AY321118 AY338174 AY338175 AY348314 AP006557 AP006558 AP006559 AP006560 AP006561 AY427439 AY322205 AY322206 AY322207 AY463059 AY269391 AY268049
Proteomes
PRIDE
Pfam
Interpro
IPR002589
Macro_dom
IPR032592 NAR_dom
IPR038400 Nsp3_coronavir_sf
IPR036333 NSP10_sf
IPR038083 R1a/1ab
IPR024375 Nsp3_coronavir
IPR009466 NSP11
IPR014822 NSP9
IPR032505 Corona_NSP4_C
IPR008740 Peptidase_C30
IPR029063 SAM-dependent_MTases
IPR009469 RNA_pol_N_coronovir
IPR027352 CV_ZBD
IPR022733 Nsp3_PL2pro
IPR024358 SARS-CoV_Nsp3_N
IPR018995 RNA_synth_NSP10_coronavirus
IPR009003 Peptidase_S1_PA
IPR042515 Nsp15_N
IPR027351 (+)RNA_virus_helicase_core_dom
IPR013016 Peptidase_C30/C16
IPR036499 NSP9_sf
IPR038166 Nsp3_PL2pro_sf
IPR037230 NSP8_sf
IPR038123 NSP4_C_sf
IPR009461 Coronavirus_NSP16
IPR014828 NSP7
IPR038030 NSP1_sf
IPR042570 NAR_sf
IPR014829 NSP8
IPR027417 P-loop_NTPase
IPR021590 NSP1
IPR014827 Viral_protease
IPR007094 RNA-dir_pol_PSvirus
IPR037204 NSP7_sf
IPR037227 EndoU-like
IPR032592 NAR_dom
IPR038400 Nsp3_coronavir_sf
IPR036333 NSP10_sf
IPR038083 R1a/1ab
IPR024375 Nsp3_coronavir
IPR009466 NSP11
IPR014822 NSP9
IPR032505 Corona_NSP4_C
IPR008740 Peptidase_C30
IPR029063 SAM-dependent_MTases
IPR009469 RNA_pol_N_coronovir
IPR027352 CV_ZBD
IPR022733 Nsp3_PL2pro
IPR024358 SARS-CoV_Nsp3_N
IPR018995 RNA_synth_NSP10_coronavirus
IPR009003 Peptidase_S1_PA
IPR042515 Nsp15_N
IPR027351 (+)RNA_virus_helicase_core_dom
IPR013016 Peptidase_C30/C16
IPR036499 NSP9_sf
IPR038166 Nsp3_PL2pro_sf
IPR037230 NSP8_sf
IPR038123 NSP4_C_sf
IPR009461 Coronavirus_NSP16
IPR014828 NSP7
IPR038030 NSP1_sf
IPR042570 NAR_sf
IPR014829 NSP8
IPR027417 P-loop_NTPase
IPR021590 NSP1
IPR014827 Viral_protease
IPR007094 RNA-dir_pol_PSvirus
IPR037204 NSP7_sf
IPR037227 EndoU-like
SUPFAM
Gene 3D
ProteinModelPortal
PDB
6NUS
E-value=0,
Score=5011
Ontologies
GO
GO:0039690 P:positive stranded viral RNA replication
GO:0004483 F:mRNA (nucleoside-2'-O-)-methyltransferase activity
GO:0008270 F:zinc ion binding
GO:0004519 F:endonuclease activity
GO:0000175 F:3'-5'-exoribonuclease activity
GO:0003678 F:DNA helicase activity
GO:0039502 P:suppression by virus of host type I interferon-mediated signaling pathway
GO:0008168 F:methyltransferase activity
GO:1990380 F:Lys48-specific deubiquitinase activity
GO:0090503 P:RNA phosphodiester bond hydrolysis, exonucleolytic
GO:0039548 P:suppression by virus of host IRF3 activity
GO:0019082 P:viral protein processing
GO:0039520 P:induction by virus of host autophagy
GO:0039648 P:modulation by virus of host protein ubiquitination
GO:0039694 P:viral RNA genome replication
GO:0042802 F:identical protein binding
GO:0039604 P:suppression by virus of host translation
GO:0032259 P:methylation
GO:0039714 C:cytoplasmic viral factory
GO:0044172 C:host cell endoplasmic reticulum-Golgi intermediate compartment
GO:0004482 F:mRNA (guanine-N7-)-methyltransferase activity
GO:0004197 F:cysteine-type endopeptidase activity
GO:2000158 P:positive regulation of ubiquitin-specific protease activity
GO:0044220 C:host cell perinuclear region of cytoplasm
GO:0003725 F:double-stranded RNA binding
GO:0016021 C:integral component of membrane
GO:0006370 P:7-methylguanosine mRNA capping
GO:0003724 F:RNA helicase activity
GO:0005524 F:ATP binding
GO:0036459 F:thiol-dependent ubiquitinyl hydrolase activity
GO:0003723 F:RNA binding
GO:0019083 P:viral transcription
GO:0080009 P:mRNA methylation
GO:0003968 F:RNA-directed 5'-3' RNA polymerase activity
GO:0039595 P:induction by virus of catabolism of host mRNA
GO:0003727 F:single-stranded RNA binding
GO:0004386 F:helicase activity
GO:0039579 P:suppression by virus of host ISG15 activity
GO:0033644 C:host cell membrane
GO:0039519 P:modulation by virus of host autophagy
GO:0001172 P:transcription, RNA-templated
GO:0039644 P:suppression by virus of host NF-kappaB transcription factor activity
GO:0008242 F:omega peptidase activity
GO:0006351 P:transcription, DNA-templated
GO:0004483 F:mRNA (nucleoside-2'-O-)-methyltransferase activity
GO:0008270 F:zinc ion binding
GO:0004519 F:endonuclease activity
GO:0000175 F:3'-5'-exoribonuclease activity
GO:0003678 F:DNA helicase activity
GO:0039502 P:suppression by virus of host type I interferon-mediated signaling pathway
GO:0008168 F:methyltransferase activity
GO:1990380 F:Lys48-specific deubiquitinase activity
GO:0090503 P:RNA phosphodiester bond hydrolysis, exonucleolytic
GO:0039548 P:suppression by virus of host IRF3 activity
GO:0019082 P:viral protein processing
GO:0039520 P:induction by virus of host autophagy
GO:0039648 P:modulation by virus of host protein ubiquitination
GO:0039694 P:viral RNA genome replication
GO:0042802 F:identical protein binding
GO:0039604 P:suppression by virus of host translation
GO:0032259 P:methylation
GO:0039714 C:cytoplasmic viral factory
GO:0044172 C:host cell endoplasmic reticulum-Golgi intermediate compartment
GO:0004482 F:mRNA (guanine-N7-)-methyltransferase activity
GO:0004197 F:cysteine-type endopeptidase activity
GO:2000158 P:positive regulation of ubiquitin-specific protease activity
GO:0044220 C:host cell perinuclear region of cytoplasm
GO:0003725 F:double-stranded RNA binding
GO:0016021 C:integral component of membrane
GO:0006370 P:7-methylguanosine mRNA capping
GO:0003724 F:RNA helicase activity
GO:0005524 F:ATP binding
GO:0036459 F:thiol-dependent ubiquitinyl hydrolase activity
GO:0003723 F:RNA binding
GO:0019083 P:viral transcription
GO:0080009 P:mRNA methylation
GO:0003968 F:RNA-directed 5'-3' RNA polymerase activity
GO:0039595 P:induction by virus of catabolism of host mRNA
GO:0003727 F:single-stranded RNA binding
GO:0004386 F:helicase activity
GO:0039579 P:suppression by virus of host ISG15 activity
GO:0033644 C:host cell membrane
GO:0039519 P:modulation by virus of host autophagy
GO:0001172 P:transcription, RNA-templated
GO:0039644 P:suppression by virus of host NF-kappaB transcription factor activity
GO:0008242 F:omega peptidase activity
GO:0006351 P:transcription, DNA-templated
Subcellular Location
From MSLVP
Capsid
From Uniprot
Host membrane
Host cytoplasm
nsp7, nsp8, nsp9 and nsp10 are localized in cytoplasmic foci, largely perinuclear. Late in infection, they merge into confluent complexes (By similarity). With evidence from 32 publications.
Host endoplasmic reticulum-Golgi intermediate compartment The helicase interacts with the N protein in membranous complexes and colocalizes with sites of synthesis of new viral RNA. With evidence from 32 publications.
Host cytoplasm
nsp7, nsp8, nsp9 and nsp10 are localized in cytoplasmic foci, largely perinuclear. Late in infection, they merge into confluent complexes (By similarity). With evidence from 32 publications.
Host endoplasmic reticulum-Golgi intermediate compartment The helicase interacts with the N protein in membranous complexes and colocalizes with sites of synthesis of new viral RNA. With evidence from 32 publications.
Topology
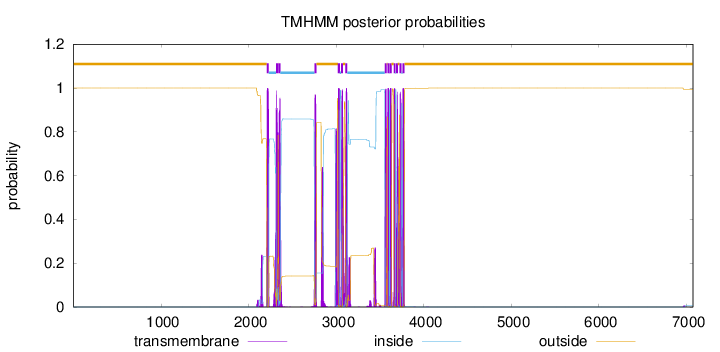
Length:
7073
Number of predicted TMHs:
14
Exp number of AAs in TMHs:
364.839299999997
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00012
outside
1 - 2208
TMhelix
2209 - 2231
inside
2232 - 2313
TMhelix
2314 - 2336
outside
2337 - 2340
TMhelix
2341 - 2363
inside
2364 - 2752
TMhelix
2753 - 2775
outside
2776 - 3019
TMhelix
3020 - 3042
inside
3043 - 3054
TMhelix
3055 - 3077
outside
3078 - 3104
TMhelix
3105 - 3127
inside
3128 - 3554
TMhelix
3555 - 3577
outside
3578 - 3586
TMhelix
3587 - 3606
inside
3607 - 3610
TMhelix
3611 - 3633
outside
3634 - 3658
TMhelix
3659 - 3678
inside
3679 - 3684
TMhelix
3685 - 3704
outside
3705 - 3723
TMhelix
3724 - 3746
inside
3747 - 3758
TMhelix
3759 - 3781
outside
3782 - 7073
Population Genetic Test Statistics
Pi
0.4723032
Theta
0.508211
Tajima's D
-0.2892304
CLR
4.64822
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 |
Protein |
QHD43415.1 |
|
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 |
Protein |
NP_828849.2 |
|
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 |
Protein |
YP_003858583.1 |
|
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.