Strain
CHINA_HS_2004_NC_006577
(Region: China: Hong Kong; Strain: Human coronavirus HKU1, complete genome.; Date: 2004)
Gene
hemagglutinin-esterase glycoprotein
Description
Annotated in NCBI,
hemagglutinin-esterase glycoprotein
Location
GenBank Accession
Full name
Hemagglutinin-esterase
Alternative Name
E3 glycoprotein
Sequence
CDS
ATGTTAATTATTTTTTTATTTTTTTATTTCTGTTATGGTTTTAATGAACCTCTTAATGTTGTGTCTCATTTAAACCATGACTGGTTTTTATTTGGTGATAGTCGTTCTGATTGTAACCATATTAATAATTTAAAAATTAAAAATTTTGATTATTTGGATATTCACCCTAGTTTGTGCAACAATGGTAAGATTTCATCTAGTGCCGGTGATTCTATTTTTAAGAGTTTTCATTTCACTCGATTTTATAATTACACTGGCGAAGGTGATCAAATTATTTTTTATGAGGGTGTTAATTTTAATCCTTATCATAGATTTAAGTGTTTTCCTAATGGTAGTAATGATGTATGGCTTCTTAACAAGGTAAGATTTTATCGTGCCTTATATTCTAATATGGCCTTTTTTCGTTATCTTACTTTTGTTGATATTCCTTATAATGTTTCTCTTTCTAAGTTTAATTCTTGTAAAAGTGATATTTTATCACTTAACAATCCTATTTTTATTAATTATTCTAAGGAAGTTTATTTTACTTTATTAGGTTGTTCTCTTTATTTAGTACCGCTTTGCCTTTTTAAATCTAACTTTAGTCAGTACTATTATAACATAGATACTGGCTCTGTTTATGGTTTTTCTAATGTTGTTTATCCTGATTTAGACTGTATTTATATTTCTCTTAAACCAGGTTCTTATAAAGTTTCCACCACTGCACCTTTTTTATCCTTACCTACTAAAGCTCTCTGTTTTGATAAATCTAAACAATTTGTACCTGTACAGGTTGTTGATTCTAGATGGAACAACGAGCGTGCCTCAGATATTTCTTTATCTGTTGCATGTCAATTGCCATATTGTTATTTTCGCAATTCTTCTGCTAATTATGTTGGCAAGTATGATATTAACCACGGTGATAGTGGTTTTATTTCTATTTTATCTGGTCTTTTATATAATGTTTCTTGTATTTCATATTATGGTGTATTTTTATATGATAATTTTACATCCATTTGGCCCTATTATTCTTTTGGTAGGTGTCCTACATCTTCTATTATTAAACATCCAATTTGTGTTTATGATTTTTTGCCTATTATTTTACAAGGTATTTTATTATGTTTAGCTTTACTTTTTGTTGTTTTTCTATTATTTTTGTTATATAACGATAAATCTCATTAA
Protein
MLIIFLFFYFCYGFNEPLNVVSHLNHDWFLFGDSRSDCNHINNLKIKNFDYLDIHPSLCNNGKISSSAGDSIFKSFHFTRFYNYTGEGDQIIFYEGVNFNPYHRFKCFPNGSNDVWLLNKVRFYRALYSNMAFFRYLTFVDIPYNVSLSKFNSCKSDILSLNNPIFINYSKEVYFTLLGCSLYLVPLCLFKSNFSQYYYNIDTGSVYGFSNVVYPDLDCIYISLKPGSYKVSTTAPFLSLPTKALCFDKSKQFVPVQVVDSRWNNERASDISLSVACQLPYCYFRNSSANYVGKYDINHGDSGFISILSGLLYNVSCISYYGVFLYDNFTSIWPYYSFGRCPTSSIIKHPICVYDFLPIILQGILLCLALLFVVFLLFLLYNDKSH
Summary
Function
Structural protein that makes short spikes at the surface of the virus. Contains receptor binding and receptor-destroying activities. Mediates de-O-acetylation of N-acetyl-4-O-acetylneuraminic acid, which is probably the receptor determinant recognized by the virus on the surface of erythrocytes and susceptible cells. This receptor-destroying activity is important for virus release as it probably helps preventing self-aggregation and ensures the efficient spread of the progeny virus from cell to cell. May serve as a secondary viral attachment protein for initiating infection, the spike protein being the major one. May become a target for both the humoral and the cellular branches of the immune system.
Catalytic Activity
H2O + N-acetyl-4-O-acetylneuraminate = acetate + H(+) + N-acetylneuraminate
H2O + N-acetyl-9-O-acetylneuraminate = acetate + H(+) + N-acetylneuraminate
H2O + N-acetyl-9-O-acetylneuraminate = acetate + H(+) + N-acetylneuraminate
Subunit
Homodimer; disulfide-linked. Forms a complex with the M protein in the pre-Golgi. Associates then with S-M complex to form a ternary complex S-M-HE.
Miscellaneous
Isolate N1 belongs to genotype A.
Isolate N2 belongs to genotype B.
Isolate N5 belongs to genotype C. Genotype C probably arose from recombination between genotypes A and B.
Isolate N2 belongs to genotype B.
Isolate N5 belongs to genotype C. Genotype C probably arose from recombination between genotypes A and B.
Similarity
Belongs to the influenza type C/coronaviruses hemagglutinin-esterase family.
Keywords
Disulfide bond
Glycoprotein
Hemagglutinin
Host cell membrane
Host membrane
Hydrolase
Membrane
Signal
Transmembrane
Transmembrane helix
Viral envelope protein
Virion
3D-structure
Reference proteome
Feature
chain Hemagglutinin-esterase
Uniprot
Q0ZJB8
Q5MQD1
S5ZBQ3
E0YJ43
Q0ZJC6
A0A140H1H0
+ More
Q0ZJ86 A0A1Y0EV02 A0A3Q9JNH8 Q0ZJB0 A0A3G2KXK9 Q14EB1 Q0ZJI2 S5YGE4 Q0ZJA2 Q0ZME8 Q0ZJ94 Q0ZJL4 A0A2H4MWZ5 A0A3G3NHQ1 A0A3G3NHB2 A0A2H4MZ25 A0A096XNG5 A0A2H4N008 U3M7K0 A0A2H4MWY2 A0A482K8Y4 A0A1S6KXR2 G5CJB0 U3M7J8 D4QFK0 U3M823 A0A0K0L8U2 Q5ICX3 O92367 U3M7I4 A0A0K0L8G7 A0A0K0KP40 A0A0B4Q7Z8 A0A5B9MYI1 A0A0B4Q9K4 U3M809 A0A2R3TWT8 U3M7J6 A0A1W6DZQ9 U3M7Y9 U3M821 A0A0K0L8E4 A0A482K8X4 A0A0K0KPJ7 A0A0K0KPP1 B8RIQ6 A0A0K0L8T5 A0A0K0L8U3 A0A2D1QW73 U3M829 U3M7J2 A0A1W6DZR7 A0A0B4Q7Z5 A0A0B4Q8M6 A0A0K0KP42 D0EY72 A0A0K0KPE0 A0A0K0KPN9 A0A411D561 D0EY92 Q4VIE5 A0A1V0E0X2 G5CJB8 P30215 A0A0K0KP17 A0A385H7F6 A0A0K0KQ35 Q4VID6 A0A0K0KQ45 U3M7Z3 A0A0K0KPN4 A0A482KB11 A0A0K0KPL2 A0A0K0L9C4 A0A0K0KPI4 A3E2F1 A0A1S5VGE8 B9TXU2 A0A0A0YP42 D0EY82 A0A0K0L8A5 A0A0K0KQ48 A0A0K0L8E7 A0A482KA41 A0A0K0KP12 A0A0K0KPI9 A0A482KDP9 A0A0K0L8A6 A0A140E064 A0A385H745 A0A482K8T1 A0A0K0KQ19 A0A385H5X6 A0A482KEQ8
Q0ZJ86 A0A1Y0EV02 A0A3Q9JNH8 Q0ZJB0 A0A3G2KXK9 Q14EB1 Q0ZJI2 S5YGE4 Q0ZJA2 Q0ZME8 Q0ZJ94 Q0ZJL4 A0A2H4MWZ5 A0A3G3NHQ1 A0A3G3NHB2 A0A2H4MZ25 A0A096XNG5 A0A2H4N008 U3M7K0 A0A2H4MWY2 A0A482K8Y4 A0A1S6KXR2 G5CJB0 U3M7J8 D4QFK0 U3M823 A0A0K0L8U2 Q5ICX3 O92367 U3M7I4 A0A0K0L8G7 A0A0K0KP40 A0A0B4Q7Z8 A0A5B9MYI1 A0A0B4Q9K4 U3M809 A0A2R3TWT8 U3M7J6 A0A1W6DZQ9 U3M7Y9 U3M821 A0A0K0L8E4 A0A482K8X4 A0A0K0KPJ7 A0A0K0KPP1 B8RIQ6 A0A0K0L8T5 A0A0K0L8U3 A0A2D1QW73 U3M829 U3M7J2 A0A1W6DZR7 A0A0B4Q7Z5 A0A0B4Q8M6 A0A0K0KP42 D0EY72 A0A0K0KPE0 A0A0K0KPN9 A0A411D561 D0EY92 Q4VIE5 A0A1V0E0X2 G5CJB8 P30215 A0A0K0KP17 A0A385H7F6 A0A0K0KQ35 Q4VID6 A0A0K0KQ45 U3M7Z3 A0A0K0KPN4 A0A482KB11 A0A0K0KPL2 A0A0K0L9C4 A0A0K0KPI4 A3E2F1 A0A1S5VGE8 B9TXU2 A0A0A0YP42 D0EY82 A0A0K0L8A5 A0A0K0KQ48 A0A0K0L8E7 A0A482KA41 A0A0K0KP12 A0A0K0KPI9 A0A482KDP9 A0A0K0L8A6 A0A140E064 A0A385H745 A0A482K8T1 A0A0K0KQ19 A0A385H5X6 A0A482KEQ8
EC Number
3.1.1.53
Pubmed
EMBL
DQ415900
DQ415901
DQ415903
DQ415904
DQ415905
DQ415906
+ More
DQ415907 DQ415908 DQ415910 KF430198 KF430203 KF686344 ABD75528.1 ABD75536.1 ABD75552.1 ABD75560.1 ABD75568.1 ABD75576.1 ABD75584.1 ABD75592.1 ABD75608.1 AGT17762.1 AGT17776.1 AGW27871.1 AY597011 KF430201 KF686340 KF686341 KF686342 KF686343 KF686346 KY674941 KY674942 KY674943 AGT17768.1 AGW27835.1 AGW27844.1 AGW27853.1 AGW27862.1 AGW27880.1 ARB07598.1 ARB07607.1 ARB07616.1 HM034837 ADN03338.1 DQ415909 ABD75600.1 KT779555 KT779556 AMN88685.1 AMN88693.1 DQ415896 DQ415914 ABD75496.1 ABD75640.1 KY983584 ARU07576.1 MK167038 AZS52617.1 DQ415911 ABD75616.1 MH940245 AYN64560.1 AY884001 DQ415902 ABD75544.1 KF430197 KY674921 AGT17757.1 ARB07437.1 DQ415897 DQ415899 DQ415912 ABD75504.1 ABD75520.1 ABD75624.1 DQ339101 DQ415913 ABD75632.1 DQ415898 ABD75512.1 KY370049 ATP66761.1 MH687968 AYR18598.1 MH687970 AYR18615.1 KY370051 ATP66772.1 KF294370 AID16648.1 KY370048 ATP66755.1 KF530071 KF530076 KF530088 KF530091 AGT51520.1 AGT51569.1 AGT51689.1 AGT51719.1 KY370043 ATP66726.1 MK327281 QBP84758.1 KX432213 AQT26497.1 JN129834 KF572664 KF572671 KF572672 KF923899 KF923900 AEN19357.1 AIL49344.1 AIL49351.1 AIL49352.1 AIV41853.1 AIV41859.1 KF530068 KF530070 KF530084 KF530098 AGT51490.1 AGT51510.1 AGT51649.1 AGT51789.1 AB551247 BAJ04695.1 KF530096 AGT51769.1 KF572702 KF923923 AIL49382.1 AIV41998.1 AY771998 AAW47239.1 AF091734 KF530063 AGT51440.1 KF923905 AIV41890.1 KF572663 AIL49343.1 KF572665 KF572666 KF923896 AIL49345.1 AIL49346.1 AIV41835.1 MN306043 QEG03775.1 KF572658 KF572661 KF923887 KF923888 AIL49338.1 AIL49341.1 AIV41782.1 AIV41788.1 KF530082 KF530089 KF530094 AGT51629.1 AGT51699.1 AGT51749.1 MF314143 AVQ05263.1 KF530069 AGT51500.1 KY967360 MN026165 ARK08669.1 QDH43725.1 KF530079 AGT51599.1 KF530095 AGT51759.1 KF923894 AIV41824.1 MK303624 QBP84748.1 KF572655 KF923886 AIL49335.1 AIV41776.1 KF572627 KF572705 AIL49307.1 AIL49385.1 EU999954 ACL12995.1 KF923925 AIV42010.1 KF923922 AIV41992.1 KY014282 ATP16766.1 KF530099 AGT51799.1 KF530067 AGT51480.1 KY967356 ARK08634.1 KF572628 KF572630 KF572631 KF572634 KF572635 KF572636 KF572637 KF572647 KF572648 KF572649 KF572650 KF572651 KF572673 KF572674 KF572675 KF572676 KF572677 KF572678 KF572679 KF572680 KF572681 KF572682 KF572683 KF572684 KF572685 KF572686 KF572687 KF572688 KF572689 KF572690 KF572691 KF572692 KF572693 KF572694 KF572696 KF572697 KF572698 KF572699 KF572701 KF572703 KF572704 KF572706 KF572707 KF923890 KF923891 KF923892 KF923901 KF923907 KF923908 KF923909 KF923910 KF923911 KF923912 KF923913 KF923914 KF923915 KF923916 KF923917 KF923918 KF923919 KF923920 KF923921 KF923924 KX344031 KY674917 KY674918 KY554972 KY554973 AIL49308.1 AIL49310.1 AIL49311.1 AIL49314.1 AIL49315.1 AIL49316.1 AIL49317.1 AIL49327.1 AIL49328.1 AIL49329.1 AIL49330.1 AIL49331.1 AIL49353.1 AIL49354.1 AIL49355.1 AIL49356.1 AIL49357.1 AIL49358.1 AIL49359.1 AIL49360.1 AIL49361.1 AIL49362.1 AIL49363.1 AIL49364.1 AIL49365.1 AIL49366.1 AIL49367.1 AIL49368.1 AIL49369.1 AIL49370.1 AIL49371.1 AIL49372.1 AIL49373.1 AIL49374.1 AIL49376.1 AIL49377.1 AIL49378.1 AIL49379.1 AIL49381.1 AIL49383.1 AIL49384.1 AIL49386.1 AIL49387.1 AIV41800.1 AIV41806.1 AIV41812.1 AIV41865.1 AIV41902.1 AIV41908.1 AIV41914.1 AIV41920.1 AIV41926.1 AIV41932.1 AIV41938.1 AIV41944.1 AIV41950.1 AIV41956.1 AIV41962.1 AIV41968.1 AIV41974.1 AIV41980.1 AIV41986.1 AIV42004.1 AOL02452.1 ARB07413.1 ARB07419.1 ARE29996.1 ARE30005.1 KF923906 AIV41896.1 KF572669 AIL49349.1 GQ918141 ACX46839.1 KF572656 AIL49336.1 KF572700 AIL49380.1 MH249786 QAY30019.1 GQ918143 ACX46859.1 AY903459 AAX85668.1 KY674920 KY554974 KY554975 ARB07432.1 ARE30014.1 ARE30023.1 JN129835 AEN19365.1 M76373 AY391777 AY585228 AY585229 KF572644 AIL49324.1 MG197712 AXX83314.1 KF572652 AIL49332.1 AY903460 AAX85677.1 KF572662 AIL49342.1 KF530081 AGT51619.1 KF572695 AIL49375.1 MK303623 QBP84739.1 KF572638 KF572641 KF572646 KF572670 KF923897 KF923903 KX538965 KX538967 KX538969 KX538971 KX538973 KX538974 KX538977 AIL49318.1 AIL49321.1 AIL49326.1 AIL49350.1 AIV41841.1 AIV41878.1 AQN78663.1 AQN78679.1 AQN78695.1 AQN78711.1 AQN78727.1 AQN78735.1 AQN78759.1 KF572668 KF923898 AIL49348.1 AIV41847.1 KF572640 KF923902 KX538970 KX538978 KX538979 MG197710 MG197713 MG197715 MG197716 MG197717 MG197722 MK303620 AIL49320.1 AIV41872.1 AQN78703.1 AQN78767.1 AQN78775.1 AXX83302.1 AXX83320.1 AXX83332.1 AXX83338.1 AXX83344.1 AXX83374.1 QBP84712.1 DQ682406 ABG78747.1 KX538968 AQN78687.1 EU401979 ACB30199.1 KF572633 KF572643 KF572660 KJ958219 AIL49313.1 AIL49323.1 AIL49340.1 AIX09806.1 GQ918142 JX860640 ACX46849.1 AFW97359.1 KF572653 KF572657 KF923889 AIL49333.1 AIL49337.1 AIV41794.1 KF572667 AIL49347.1 KF572629 KF923895 AIL49309.1 AIV41829.1 MK303625 QBQ01838.1 KF572639 KF572642 KF923904 KX538964 KX538966 KX538972 AIL49319.1 AIL49322.1 AIV41884.1 AQN78655.1 AQN78671.1 AQN78719.1 KF572645 AIL49325.1 MK303622 QBP84730.1 KF923893 AIV41818.1 KU131570 AMK59676.1 MG197711 MG197718 MG197720 MG197721 AXX83308.1 AXX83350.1 AXX83362.1 AXX83368.1 MK303621 QBP84721.1 KF572632 AIL49312.1 MG197709 AXX83296.1 MK303619 QBP84703.1
DQ415907 DQ415908 DQ415910 KF430198 KF430203 KF686344 ABD75528.1 ABD75536.1 ABD75552.1 ABD75560.1 ABD75568.1 ABD75576.1 ABD75584.1 ABD75592.1 ABD75608.1 AGT17762.1 AGT17776.1 AGW27871.1 AY597011 KF430201 KF686340 KF686341 KF686342 KF686343 KF686346 KY674941 KY674942 KY674943 AGT17768.1 AGW27835.1 AGW27844.1 AGW27853.1 AGW27862.1 AGW27880.1 ARB07598.1 ARB07607.1 ARB07616.1 HM034837 ADN03338.1 DQ415909 ABD75600.1 KT779555 KT779556 AMN88685.1 AMN88693.1 DQ415896 DQ415914 ABD75496.1 ABD75640.1 KY983584 ARU07576.1 MK167038 AZS52617.1 DQ415911 ABD75616.1 MH940245 AYN64560.1 AY884001 DQ415902 ABD75544.1 KF430197 KY674921 AGT17757.1 ARB07437.1 DQ415897 DQ415899 DQ415912 ABD75504.1 ABD75520.1 ABD75624.1 DQ339101 DQ415913 ABD75632.1 DQ415898 ABD75512.1 KY370049 ATP66761.1 MH687968 AYR18598.1 MH687970 AYR18615.1 KY370051 ATP66772.1 KF294370 AID16648.1 KY370048 ATP66755.1 KF530071 KF530076 KF530088 KF530091 AGT51520.1 AGT51569.1 AGT51689.1 AGT51719.1 KY370043 ATP66726.1 MK327281 QBP84758.1 KX432213 AQT26497.1 JN129834 KF572664 KF572671 KF572672 KF923899 KF923900 AEN19357.1 AIL49344.1 AIL49351.1 AIL49352.1 AIV41853.1 AIV41859.1 KF530068 KF530070 KF530084 KF530098 AGT51490.1 AGT51510.1 AGT51649.1 AGT51789.1 AB551247 BAJ04695.1 KF530096 AGT51769.1 KF572702 KF923923 AIL49382.1 AIV41998.1 AY771998 AAW47239.1 AF091734 KF530063 AGT51440.1 KF923905 AIV41890.1 KF572663 AIL49343.1 KF572665 KF572666 KF923896 AIL49345.1 AIL49346.1 AIV41835.1 MN306043 QEG03775.1 KF572658 KF572661 KF923887 KF923888 AIL49338.1 AIL49341.1 AIV41782.1 AIV41788.1 KF530082 KF530089 KF530094 AGT51629.1 AGT51699.1 AGT51749.1 MF314143 AVQ05263.1 KF530069 AGT51500.1 KY967360 MN026165 ARK08669.1 QDH43725.1 KF530079 AGT51599.1 KF530095 AGT51759.1 KF923894 AIV41824.1 MK303624 QBP84748.1 KF572655 KF923886 AIL49335.1 AIV41776.1 KF572627 KF572705 AIL49307.1 AIL49385.1 EU999954 ACL12995.1 KF923925 AIV42010.1 KF923922 AIV41992.1 KY014282 ATP16766.1 KF530099 AGT51799.1 KF530067 AGT51480.1 KY967356 ARK08634.1 KF572628 KF572630 KF572631 KF572634 KF572635 KF572636 KF572637 KF572647 KF572648 KF572649 KF572650 KF572651 KF572673 KF572674 KF572675 KF572676 KF572677 KF572678 KF572679 KF572680 KF572681 KF572682 KF572683 KF572684 KF572685 KF572686 KF572687 KF572688 KF572689 KF572690 KF572691 KF572692 KF572693 KF572694 KF572696 KF572697 KF572698 KF572699 KF572701 KF572703 KF572704 KF572706 KF572707 KF923890 KF923891 KF923892 KF923901 KF923907 KF923908 KF923909 KF923910 KF923911 KF923912 KF923913 KF923914 KF923915 KF923916 KF923917 KF923918 KF923919 KF923920 KF923921 KF923924 KX344031 KY674917 KY674918 KY554972 KY554973 AIL49308.1 AIL49310.1 AIL49311.1 AIL49314.1 AIL49315.1 AIL49316.1 AIL49317.1 AIL49327.1 AIL49328.1 AIL49329.1 AIL49330.1 AIL49331.1 AIL49353.1 AIL49354.1 AIL49355.1 AIL49356.1 AIL49357.1 AIL49358.1 AIL49359.1 AIL49360.1 AIL49361.1 AIL49362.1 AIL49363.1 AIL49364.1 AIL49365.1 AIL49366.1 AIL49367.1 AIL49368.1 AIL49369.1 AIL49370.1 AIL49371.1 AIL49372.1 AIL49373.1 AIL49374.1 AIL49376.1 AIL49377.1 AIL49378.1 AIL49379.1 AIL49381.1 AIL49383.1 AIL49384.1 AIL49386.1 AIL49387.1 AIV41800.1 AIV41806.1 AIV41812.1 AIV41865.1 AIV41902.1 AIV41908.1 AIV41914.1 AIV41920.1 AIV41926.1 AIV41932.1 AIV41938.1 AIV41944.1 AIV41950.1 AIV41956.1 AIV41962.1 AIV41968.1 AIV41974.1 AIV41980.1 AIV41986.1 AIV42004.1 AOL02452.1 ARB07413.1 ARB07419.1 ARE29996.1 ARE30005.1 KF923906 AIV41896.1 KF572669 AIL49349.1 GQ918141 ACX46839.1 KF572656 AIL49336.1 KF572700 AIL49380.1 MH249786 QAY30019.1 GQ918143 ACX46859.1 AY903459 AAX85668.1 KY674920 KY554974 KY554975 ARB07432.1 ARE30014.1 ARE30023.1 JN129835 AEN19365.1 M76373 AY391777 AY585228 AY585229 KF572644 AIL49324.1 MG197712 AXX83314.1 KF572652 AIL49332.1 AY903460 AAX85677.1 KF572662 AIL49342.1 KF530081 AGT51619.1 KF572695 AIL49375.1 MK303623 QBP84739.1 KF572638 KF572641 KF572646 KF572670 KF923897 KF923903 KX538965 KX538967 KX538969 KX538971 KX538973 KX538974 KX538977 AIL49318.1 AIL49321.1 AIL49326.1 AIL49350.1 AIV41841.1 AIV41878.1 AQN78663.1 AQN78679.1 AQN78695.1 AQN78711.1 AQN78727.1 AQN78735.1 AQN78759.1 KF572668 KF923898 AIL49348.1 AIV41847.1 KF572640 KF923902 KX538970 KX538978 KX538979 MG197710 MG197713 MG197715 MG197716 MG197717 MG197722 MK303620 AIL49320.1 AIV41872.1 AQN78703.1 AQN78767.1 AQN78775.1 AXX83302.1 AXX83320.1 AXX83332.1 AXX83338.1 AXX83344.1 AXX83374.1 QBP84712.1 DQ682406 ABG78747.1 KX538968 AQN78687.1 EU401979 ACB30199.1 KF572633 KF572643 KF572660 KJ958219 AIL49313.1 AIL49323.1 AIL49340.1 AIX09806.1 GQ918142 JX860640 ACX46849.1 AFW97359.1 KF572653 KF572657 KF923889 AIL49333.1 AIL49337.1 AIV41794.1 KF572667 AIL49347.1 KF572629 KF923895 AIL49309.1 AIV41829.1 MK303625 QBQ01838.1 KF572639 KF572642 KF923904 KX538964 KX538966 KX538972 AIL49319.1 AIL49322.1 AIV41884.1 AQN78655.1 AQN78671.1 AQN78719.1 KF572645 AIL49325.1 MK303622 QBP84730.1 KF923893 AIV41818.1 KU131570 AMK59676.1 MG197711 MG197718 MG197720 MG197721 AXX83308.1 AXX83350.1 AXX83362.1 AXX83368.1 MK303621 QBP84721.1 KF572632 AIL49312.1 MG197709 AXX83296.1 MK303619 QBP84703.1
Proteomes
UP000102058
UP000103177
UP000107950
UP000120443
UP000123677
UP000127413
+ More
UP000147573 UP000148743 UP000164754 UP000168247 UP000008170 UP000098016 UP000098607 UP000139452 UP000143896 UP000173725 UP000173897 UP000108760 UP000174991 UP000122230 UP000134208 UP000118444 UP000161305 UP000162514 UP000006551 UP000124867 UP000134743 UP000143869 UP000168964 UP000001985 UP000156925 UP000107997 UP000097333 UP000119922 UP000150595 UP000160021 UP000119882 UP000121854 UP000173822 UP000100004 UP000117387 UP000119762 UP000168290 UP000154289 UP000158566 UP000112072 UP000172586 UP000128148 UP000174077 UP000104994 UP000143341 UP000104124 UP000109543 UP000114034 UP000155676 UP000143194 UP000140040 UP000123825 UP000114688 UP000173555 UP000114169 UP000109321 UP000122365 UP000107048 UP000108500 UP000115343 UP000115819 UP000116223 UP000117554 UP000119058 UP000119993 UP000128782 UP000130040 UP000130067 UP000132146 UP000136483 UP000136628 UP000137532 UP000142623 UP000143152 UP000150164 UP000152870 UP000166197 UP000173306 UP000161137 UP000130586 UP000007552 UP000100580 UP000180344 UP000159995 UP000140401 UP000154332 UP000162080 UP000142982 UP000101116 UP000099221 UP000137675 UP000137844 UP000161839 UP000165756 UP000149793 UP000147591
UP000147573 UP000148743 UP000164754 UP000168247 UP000008170 UP000098016 UP000098607 UP000139452 UP000143896 UP000173725 UP000173897 UP000108760 UP000174991 UP000122230 UP000134208 UP000118444 UP000161305 UP000162514 UP000006551 UP000124867 UP000134743 UP000143869 UP000168964 UP000001985 UP000156925 UP000107997 UP000097333 UP000119922 UP000150595 UP000160021 UP000119882 UP000121854 UP000173822 UP000100004 UP000117387 UP000119762 UP000168290 UP000154289 UP000158566 UP000112072 UP000172586 UP000128148 UP000174077 UP000104994 UP000143341 UP000104124 UP000109543 UP000114034 UP000155676 UP000143194 UP000140040 UP000123825 UP000114688 UP000173555 UP000114169 UP000109321 UP000122365 UP000107048 UP000108500 UP000115343 UP000115819 UP000116223 UP000117554 UP000119058 UP000119993 UP000128782 UP000130040 UP000130067 UP000132146 UP000136483 UP000136628 UP000137532 UP000142623 UP000143152 UP000150164 UP000152870 UP000166197 UP000173306 UP000161137 UP000130586 UP000007552 UP000100580 UP000180344 UP000159995 UP000140401 UP000154332 UP000162080 UP000142982 UP000101116 UP000099221 UP000137675 UP000137844 UP000161839 UP000165756 UP000149793 UP000147591
PRIDE
Interpro
SUPFAM
SSF49818
SSF49818
ProteinModelPortal
Q0ZJB8
Q5MQD1
S5ZBQ3
E0YJ43
Q0ZJC6
A0A140H1H0
+ More
Q0ZJ86 A0A1Y0EV02 A0A3Q9JNH8 Q0ZJB0 A0A3G2KXK9 Q14EB1 Q0ZJI2 S5YGE4 Q0ZJA2 Q0ZME8 Q0ZJ94 Q0ZJL4 A0A2H4MWZ5 A0A3G3NHQ1 A0A3G3NHB2 A0A2H4MZ25 A0A096XNG5 A0A2H4N008 U3M7K0 A0A2H4MWY2 A0A482K8Y4 A0A1S6KXR2 G5CJB0 U3M7J8 D4QFK0 U3M823 A0A0K0L8U2 Q5ICX3 O92367 U3M7I4 A0A0K0L8G7 A0A0K0KP40 A0A0B4Q7Z8 A0A5B9MYI1 A0A0B4Q9K4 U3M809 A0A2R3TWT8 U3M7J6 A0A1W6DZQ9 U3M7Y9 U3M821 A0A0K0L8E4 A0A482K8X4 A0A0K0KPJ7 A0A0K0KPP1 B8RIQ6 A0A0K0L8T5 A0A0K0L8U3 A0A2D1QW73 U3M829 U3M7J2 A0A1W6DZR7 A0A0B4Q7Z5 A0A0B4Q8M6 A0A0K0KP42 D0EY72 A0A0K0KPE0 A0A0K0KPN9 A0A411D561 D0EY92 Q4VIE5 A0A1V0E0X2 G5CJB8 P30215 A0A0K0KP17 A0A385H7F6 A0A0K0KQ35 Q4VID6 A0A0K0KQ45 U3M7Z3 A0A0K0KPN4 A0A482KB11 A0A0K0KPL2 A0A0K0L9C4 A0A0K0KPI4 A3E2F1 A0A1S5VGE8 B9TXU2 A0A0A0YP42 D0EY82 A0A0K0L8A5 A0A0K0KQ48 A0A0K0L8E7 A0A482KA41 A0A0K0KP12 A0A0K0KPI9 A0A482KDP9 A0A0K0L8A6 A0A140E064 A0A385H745 A0A482K8T1 A0A0K0KQ19 A0A385H5X6 A0A482KEQ8
Q0ZJ86 A0A1Y0EV02 A0A3Q9JNH8 Q0ZJB0 A0A3G2KXK9 Q14EB1 Q0ZJI2 S5YGE4 Q0ZJA2 Q0ZME8 Q0ZJ94 Q0ZJL4 A0A2H4MWZ5 A0A3G3NHQ1 A0A3G3NHB2 A0A2H4MZ25 A0A096XNG5 A0A2H4N008 U3M7K0 A0A2H4MWY2 A0A482K8Y4 A0A1S6KXR2 G5CJB0 U3M7J8 D4QFK0 U3M823 A0A0K0L8U2 Q5ICX3 O92367 U3M7I4 A0A0K0L8G7 A0A0K0KP40 A0A0B4Q7Z8 A0A5B9MYI1 A0A0B4Q9K4 U3M809 A0A2R3TWT8 U3M7J6 A0A1W6DZQ9 U3M7Y9 U3M821 A0A0K0L8E4 A0A482K8X4 A0A0K0KPJ7 A0A0K0KPP1 B8RIQ6 A0A0K0L8T5 A0A0K0L8U3 A0A2D1QW73 U3M829 U3M7J2 A0A1W6DZR7 A0A0B4Q7Z5 A0A0B4Q8M6 A0A0K0KP42 D0EY72 A0A0K0KPE0 A0A0K0KPN9 A0A411D561 D0EY92 Q4VIE5 A0A1V0E0X2 G5CJB8 P30215 A0A0K0KP17 A0A385H7F6 A0A0K0KQ35 Q4VID6 A0A0K0KQ45 U3M7Z3 A0A0K0KPN4 A0A482KB11 A0A0K0KPL2 A0A0K0L9C4 A0A0K0KPI4 A3E2F1 A0A1S5VGE8 B9TXU2 A0A0A0YP42 D0EY82 A0A0K0L8A5 A0A0K0KQ48 A0A0K0L8E7 A0A482KA41 A0A0K0KP12 A0A0K0KPI9 A0A482KDP9 A0A0K0L8A6 A0A140E064 A0A385H745 A0A482K8T1 A0A0K0KQ19 A0A385H5X6 A0A482KEQ8
PDB
5N11
E-value=1.45112e-105,
Score=978
Ontologies
KEGG
GO
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0020002 C:host cell plasma membrane
GO:0001681 F:sialate O-acetylesterase activity
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0046789 F:host cell surface receptor binding
GO:0055036 C:virion membrane
GO:0016788 F:hydrolase activity, acting on ester bonds
GO:0019058 P:viral life cycle
GO:0020002 C:host cell plasma membrane
GO:0001681 F:sialate O-acetylesterase activity
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0046789 F:host cell surface receptor binding
GO:0055036 C:virion membrane
GO:0016788 F:hydrolase activity, acting on ester bonds
GO:0019058 P:viral life cycle
Subcellular Location
From MSLVP
Single-Pass Membrane
From Uniprot
Virion membrane
In infected cells becomes incorporated into the envelope of virions during virus assembly at the endoplasmic reticulum and cis Golgi. However, some may escape incorporation into virions and subsequently migrate to the cell surface. With evidence from 1 publications.
Host cell membrane In infected cells becomes incorporated into the envelope of virions during virus assembly at the endoplasmic reticulum and cis Golgi. However, some may escape incorporation into virions and subsequently migrate to the cell surface. With evidence from 1 publications.
Host cell membrane In infected cells becomes incorporated into the envelope of virions during virus assembly at the endoplasmic reticulum and cis Golgi. However, some may escape incorporation into virions and subsequently migrate to the cell surface. With evidence from 1 publications.
Topology
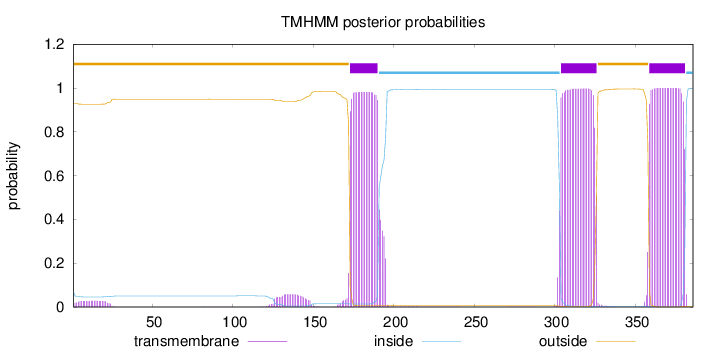
Length:
386
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
67.07051
Exp number, first 60 AAs:
0.58501
Total prob of N-in:
0.06931
outside
1 - 172
TMhelix
173 - 190
inside
191 - 303
TMhelix
304 - 326
outside
327 - 358
TMhelix
359 - 381
inside
382 - 386
Population Genetic Test Statistics
Pi
0.4978595
Theta
0.6156864
Tajima's D
-0.5637533
CLR
3.327165
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 |
Protein |
YP_173237.1 |
|
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.