Strain
CHINA_HS_2004_NC_006577
(Region: China: Hong Kong; Strain: Human coronavirus HKU1, complete genome.; Date: 2004)
Gene
spike glycoprotein
Description
Annotated in NCBI,
spike glycoprotein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGTTATTAATTATTTTTATTTTGCCTACAACATTAGCTGTTATAGGTGATTTTAATTGTACTAATTTTGCTATTAATGATTTAAACACCACAGTTCCTCGCATAAGTGAGTATGTTGTGGATGTTTCTTATGGTTTGGGTACATATTATATACTTGATCGTGTTTATTTAAATACTACTATATTATTTACTGGTTATTTCCCTAAATCTGGTGCCAATTTTAGGGATCTATCTTTAAAAGGTACTACATATTTGAGTACTCTTTGGTATCAGAAACCCTTTTTATCTGATTTTAATAATGGTATTTTTTCTAGAGTTAAGAATACTAAGTTGTATGTTAATAAAACTTTGTATAGTGAGTTTAGTACTATAGTTATAGGTAGTGTTTTTATTAACAACTCTTATACTATTGTTGTTCAACCTCATAATGGTGTTTTGGAGATTACAGCTTGTCAATACACTATGTGTGAGTATCCTCATACTATTTGTAAATCTAAAGGTAGTTCTCGTAATGAATCTTGGCATTTTGATAAATCTGAACCTTTGTGTCTGTTCAAGAAAAATTTTACTTATAATGTTTCTACAGATTGGTTGTATTTTCATTTTTATCAAGAACGTGGCACTTTTTATGCTTATTATGCTGATTCTGGCATGCCTACTACTTTTTTATTTAGTTTGTATCTTGGTACTCTTTTATCTCATTATTATGTTTTGCCTTTGACTTGTAATGCTATATCTTCTAATACTGATAATGAGACTTTACAATATTGGGTCACACCTTTGTCTAAACGCCAATATCTTCTTAAATTTGACAACCGTGGTGTTATTACTAATGCTGTTGATTGTTCTAGTAGTTTCTTTAGCGAGATTCAATGTAAAACTAAATCTTTATTACCTAATACTGGTGTTTATGACTTATCTGGTTTTACTGTTAAGCCTGTTGCAACTGTACATCGTCGTATTCCTGATTTACCTGATTGTGACATTGATAAATGGCTTAACAATTTTAATGTACCCTCACCTCTTAATTGGGAACGTAAAATTTTTTCTAATTGCAACTTTAATTTGAGTACTTTGCTTCGTTTAGTTCATACTGATTCTTTTTCTTGTAATAATTTTGATGAATCTAAGATATATGGTAGTTGTTTTAAGAGTATTGTTTTAGATAAATTTGCCATACCCAACTCCAGACGATCTGATTTGCAGTTGGGCAGTTCTGGTTTTCTGCAATCTTCTAATTATAAAATTGACACTACTTCTAGTTCTTGTCAATTGTATTATAGTTTGCCTGCAATTAATGTTACTATTAATAATTATAATCCTTCTTCTTGGAATAGAAGGTATGGTTTTAATAATTTTAATTTGAGCTCTCATAGTGTTGTTTACTCACGTTATTGTTTTTCTGTTAATAATACTTTTTGTCCTTGTGCTAAACCTTCTTTTGCTTCAAGTTGCAAGAGTCATAAACCACCTTCTGCTTCCTGTCCTATTGGTACTAATTATCGTTCTTGTGAGAGTACTACTGTACTCGACCACACTGACTGGTGTAGGTGTTCTTGTTTACCTGATCCTATAACTGCTTATGACCCTAGGTCTTGTTCTCAAAAAAAGTCTCTGGTTGGTGTTGGTGAACATTGTGCAGGGTTCGGTGTTGATGAAGAAAAGTGTGGTGTATTGGATGGATCATATAATGTTTCTTGTCTTTGTAGTACTGATGCCTTTCTAGGTTGGTCTTATGACACTTGCGTCAGTAACAACCGTTGTAATATTTTTTCTAATTTTATTTTAAATGGTATCAATAGTGGTACCACTTGTTCTAATGATTTATTGCAGCCTAATACTGAAGTTTTTACTGATGTTTGTGTTGATTACGACCTTTATGGTATTACAGGACAAGGTATTTTTAAAGAAGTTTCTGCTGTTTATTATAATAGTTGGCAAAATCTTTTGTATGATTCTAATGGCAACATTATTGGTTTTAAAGATTTTGTTACTAATAAAACATATAATATTTTCCCTTGTTATGCAGGAAGAGTTTCTGCTGCTTTTCATCAAAATGCTTCCTCTTTGGCTTTACTTTATCGTAATTTAAAATGTAGCTATGTTTTGAATAATATTTCTTTAACTACTCAGCCATATTTTGATAGTTATCTTGGTTGCGTTTTTAATGCTGATAATTTAACTGATTATTCTGTTTCTTCTTGTGCTCTTCGCATGGGTAGTGGTTTTTGTGTTGATTATAACTCACCTTCTTCTTCCTCTTCGCGTCGTAAACGTAGAAGTATTTCTGCTTCTTATCGTTTTGTTACTTTTGAACCCTTTAATGTCAGTTTTGTTAATGACAGTATTGAGTCTGTGGGTGGTCTTTATGAGATCAAAATTCCCACTAACTTTACTATAGTTGGTCAAGAGGAATTTATTCAAACTAATTCTCCTAAAGTTACTATTGATTGTTCTTTATTTGTCTGTTCTAATTATGCAGCTTGCCATGACTTATTGTCAGAGTATGGCACTTTTTGTGATAATATTAATAGTATTTTAGATGAAGTTAATGGTTTACTTGATACTACTCAATTGCATGTAGCTGATACTCTTATGCAAGGTGTCACACTTAGCTCCAATCTTAATACTAATTTGCATTTTGATGTTGATAATATTAATTTTAAATCCCTAGTTGGATGTTTAGGTCCACACTGCGGTTCTTCTTCTCGTTCTTTTTTTGAAGATTTATTGTTTGACAAAGTTAAACTTTCAGATGTTGGTTTTGTTGAAGCTTATAACAATTGTACTGGTGGTAGTGAAATTAGAGATCTTCTTTGTGTACAATCCTTTAATGGTATTAAAGTTTTGCCTCCTATTTTGTCTGAATCTCAAATTTCTGGTTACACCACAGCCGCTACTGTTGCTGCTATGTTTCCACCATGGTCAGCAGCAGCTGGCATACCATTTTCTCTTAATGTACAATATAGAATTAATGGTTTGGGTGTTACTATGGATGTTCTTAATAAAAATCAAAAGTTGATAGCTACTGCTTTTAATAATGCTCTTCTTTCTATTCAGAATGGTTTTAGTGCTACCAACTCTGCACTTGCTAAAATACAAAGTGTTGTTAATTCTAATGCTCAAGCACTTAATAGTTTGTTACAGCAATTATTTAATAAATTTGGTGCAATTAGTTCTTCTTTACAAGAAATTTTATCTCGTCTCGATGCTTTAGAGGCTCAGGTTCAGATTGATAGGCTTATTAATGGTCGTTTAACTGCTTTAAATGCTTATGTCTCTCAACAGCTTAGTGATATTTCTCTTGTAAAATTTGGTGCTGCTTTAGCTATGGAGAAGGTTAATGAGTGTGTTAAAAGTCAATCTCCTCGTATTAATTTTTGTGGTAATGGTAATCATATTTTGTCATTAGTTCAAAATGCTCCTTATGGTTTGTTGTTTATGCATTTTAGTTATAAACCTATTTCTTTTAAAACTGTTTTAGTAAGTCCTGGTTTGTGTATATCAGGTGATGTAGGTATTGCACCTAAACAAGGGTATTTTATTAAACATAATGATCATTGGATGTTCACTGGTAGTTCTTACTATTATCCTGAACCAATTTCAGATAAAAATGTTGTTTTTATGAATACTTGTTCTGTTAATTTTACTAAAGCGCCTCTTGTTTATTTGAATCATTCTGTACCAAAATTGTCTGATTTTGAATCTGAGTTATCTCATTGGTTTAAAAATCAAACATCCATTGCGCCTAATTTGACTTTAAATCTTCATACTATTAATGCTACTTTTTTAGATTTGTATTATGAGATGAATCTTATTCAAGAGTCTATTAAGTCTTTGAATAATAGTTATATCAATCTTAAAGATATAGGTACATATGAAATGTATGTAAAATGGCCTTGGTATGTTTGGCTACTAATTTCTTTTTCATTTATAATATTCCTTGTATTGCTCTTTTTTATATGTTGTTGTACTGGTTGTGGTTCTGCATGTTTTAGTAAATGTCATAATTGTTGTGATGAGTATGGTGGTCATCATGATTTTGTTATCAAAACATCTCATGATGATTAG
Protein
MLLIIFILPTTLAVIGDFNCTNFAINDLNTTVPRISEYVVDVSYGLGTYYILDRVYLNTTILFTGYFPKSGANFRDLSLKGTTYLSTLWYQKPFLSDFNNGIFSRVKNTKLYVNKTLYSEFSTIVIGSVFINNSYTIVVQPHNGVLEITACQYTMCEYPHTICKSKGSSRNESWHFDKSEPLCLFKKNFTYNVSTDWLYFHFYQERGTFYAYYADSGMPTTFLFSLYLGTLLSHYYVLPLTCNAISSNTDNETLQYWVTPLSKRQYLLKFDNRGVITNAVDCSSSFFSEIQCKTKSLLPNTGVYDLSGFTVKPVATVHRRIPDLPDCDIDKWLNNFNVPSPLNWERKIFSNCNFNLSTLLRLVHTDSFSCNNFDESKIYGSCFKSIVLDKFAIPNSRRSDLQLGSSGFLQSSNYKIDTTSSSCQLYYSLPAINVTINNYNPSSWNRRYGFNNFNLSSHSVVYSRYCFSVNNTFCPCAKPSFASSCKSHKPPSASCPIGTNYRSCESTTVLDHTDWCRCSCLPDPITAYDPRSCSQKKSLVGVGEHCAGFGVDEEKCGVLDGSYNVSCLCSTDAFLGWSYDTCVSNNRCNIFSNFILNGINSGTTCSNDLLQPNTEVFTDVCVDYDLYGITGQGIFKEVSAVYYNSWQNLLYDSNGNIIGFKDFVTNKTYNIFPCYAGRVSAAFHQNASSLALLYRNLKCSYVLNNISLTTQPYFDSYLGCVFNADNLTDYSVSSCALRMGSGFCVDYNSPSSSSSRRKRRSISASYRFVTFEPFNVSFVNDSIESVGGLYEIKIPTNFTIVGQEEFIQTNSPKVTIDCSLFVCSNYAACHDLLSEYGTFCDNINSILDEVNGLLDTTQLHVADTLMQGVTLSSNLNTNLHFDVDNINFKSLVGCLGPHCGSSSRSFFEDLLFDKVKLSDVGFVEAYNNCTGGSEIRDLLCVQSFNGIKVLPPILSESQISGYTTAATVAAMFPPWSAAAGIPFSLNVQYRINGLGVTMDVLNKNQKLIATAFNNALLSIQNGFSATNSALAKIQSVVNSNAQALNSLLQQLFNKFGAISSSLQEILSRLDALEAQVQIDRLINGRLTALNAYVSQQLSDISLVKFGAALAMEKVNECVKSQSPRINFCGNGNHILSLVQNAPYGLLFMHFSYKPISFKTVLVSPGLCISGDVGIAPKQGYFIKHNDHWMFTGSSYYYPEPISDKNVVFMNTCSVNFTKAPLVYLNHSVPKLSDFESELSHWFKNQTSIAPNLTLNLHTINATFLDLYYEMNLIQESIKSLNNSYINLKDIGTYEMYVKWPWYVWLLISFSFIIFLVLLFFICCCTGCGSACFSKCHNCCDEYGGHHDFVIKTSHDD
Summary
Function
attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S1: attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S1: attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes.
Miscellaneous
Isolate N1 belongs to genotype A.
Isolate N5 belongs to genotype C. Genotype C probably arose from recombination between genotypes A and B.
Isolate N2 belongs to genotype B.
Isolate N5 belongs to genotype C. Genotype C probably arose from recombination between genotypes A and B.
Isolate N2 belongs to genotype B.
Similarity
Belongs to the betacoronaviruses spike protein family.
Keywords
3D-structure
Coiled coil
Disulfide bond
Fusion of virus membrane with host endosomal membrane
Fusion of virus membrane with host membrane
Glycoprotein
Host cell membrane
Host membrane
Host-virus interaction
Lipoprotein
Membrane
Palmitate
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Viral penetration into host cytoplasm
Virion
Virulence
Virus entry into host cell
Feature
chain Spike glycoprotein
Uniprot
Q5MQD0
Q19U42
Q19U37
S5YGG8
Q0ZJH3
Q19U49
+ More
Q19U48 A0A224AU27 U3NAI2 A0A140H1H1 U3N9S7 Q19U47 U3N885 A0A140H1H9 E0YJ44 A0A1Y0EUZ8 S5YA28 A0A451ERM8 Q0ZME7 Q19U40 Q19U39 Q19U45 Q0ZJI1 A0A3G2KXL1 Q14EB0 Q19U46 A0A224ANC9 S5ZBP3 A0A1V0E0Z4 A0A2H4MWY0 A0A3G3NGZ3 A0A2H4MX03 A0A096XNG8 A0A2H4MZ72 A0A2H4MZ10 A0A096XNI4 A0A3G3NHC8 S4WG83 R4WAW7 I2FKH1 A0A0K2RVK5 A0A0K2RVL1 A0A3S7GYI2 B9TXV2 A0A2S0SZ25 A8R4D0 Q83331 S4WIX3 E5RPZ2 S4WLK1 S4WML5 Q6W355 A0A142D826 C6GHR0 C6GHL3 D7PJY2 S4WHI9 A0A5J6NUL2 H9BZX9 A0A5J6NRR8 S4WG91 S4WG99 B7TYG9 H9AA34 B7U2L1 B3VTS5 Q0PL39 S4WIX5 A0A346I7G9 H9AA55 S4WG79 Q2EEY3 S4WLK8 A0A481S1P4 S4WLK4 P25192 B7U2R5 B7U2N3 F1T0N1 B7U2M2 B7U2S7 S4WG94 A0A142D823 A0A142D820 E3UTE9 A4ZU41 S4WLL0 A6YD40 Q58NS2 A0A142D831 Q9QD01 C6GHP9 S4WHJ6 A0A346I7H0 A0A481S1B7 A0A2R4STR1 A0A142D818 B9TXU9 Q58NS1 A0A142D828
Q19U48 A0A224AU27 U3NAI2 A0A140H1H1 U3N9S7 Q19U47 U3N885 A0A140H1H9 E0YJ44 A0A1Y0EUZ8 S5YA28 A0A451ERM8 Q0ZME7 Q19U40 Q19U39 Q19U45 Q0ZJI1 A0A3G2KXL1 Q14EB0 Q19U46 A0A224ANC9 S5ZBP3 A0A1V0E0Z4 A0A2H4MWY0 A0A3G3NGZ3 A0A2H4MX03 A0A096XNG8 A0A2H4MZ72 A0A2H4MZ10 A0A096XNI4 A0A3G3NHC8 S4WG83 R4WAW7 I2FKH1 A0A0K2RVK5 A0A0K2RVL1 A0A3S7GYI2 B9TXV2 A0A2S0SZ25 A8R4D0 Q83331 S4WIX3 E5RPZ2 S4WLK1 S4WML5 Q6W355 A0A142D826 C6GHR0 C6GHL3 D7PJY2 S4WHI9 A0A5J6NUL2 H9BZX9 A0A5J6NRR8 S4WG91 S4WG99 B7TYG9 H9AA34 B7U2L1 B3VTS5 Q0PL39 S4WIX5 A0A346I7G9 H9AA55 S4WG79 Q2EEY3 S4WLK8 A0A481S1P4 S4WLK4 P25192 B7U2R5 B7U2N3 F1T0N1 B7U2M2 B7U2S7 S4WG94 A0A142D823 A0A142D820 E3UTE9 A4ZU41 S4WLL0 A6YD40 Q58NS2 A0A142D831 Q9QD01 C6GHP9 S4WHJ6 A0A346I7H0 A0A481S1B7 A0A2R4STR1 A0A142D818 B9TXU9 Q58NS1 A0A142D828
Pubmed
EMBL
AY597011
DQ415896
DQ415904
DQ415905
DQ415906
DQ415907
+ More
DQ415914 DQ437613 DQ437614 ABD75497.1 ABD75561.1 ABD75569.1 ABD75577.1 ABD75585.1 ABD75641.1 ABD96192.1 ABD96193.1 DQ415900 DQ415901 DQ437618 DQ437619 KF430198 ABD75529.1 ABD75537.1 ABD96197.1 ABD96198.1 AGT17763.1 KF430203 AGT17777.1 DQ415903 ABD75553.1 DQ415908 DQ437607 ABD75593.1 ABD96186.1 DQ415909 DQ437608 ABD75601.1 ABD96187.1 LC315650 BBA20983.1 KF686340 KF686341 KF686342 KF686346 KY674941 KY674942 KY674943 AGW27836.1 AGW27845.1 AGW27854.1 AGW27881.1 ARB07599.1 ARB07608.1 ARB07617.1 KT779555 AMN88686.1 KF686344 AGW27872.1 DQ415910 DQ437609 ABD75609.1 ABD96188.1 KF686343 AGW27863.1 KT779556 AMN88694.1 HM034837 ADN03339.1 KY983584 ARU07577.1 KF430201 AGT17769.1 MK167038 AZS52618.1 DQ339101 DQ415898 DQ437616 ABD75513.1 ABD96195.1 DQ415897 DQ415899 DQ415913 DQ437612 DQ437615 DQ437617 ABD75505.1 ABD75521.1 ABD75633.1 ABD96191.1 ABD96194.1 ABD96196.1 DQ415912 DQ437611 ABD75625.1 ABD96190.1 DQ415902 ABD75545.1 MH940245 AYN64561.1 AY884001 DQ415911 DQ437610 ABD75617.1 ABD96189.1 LC315651 BBA20986.1 KF430197 AGT17758.1 KY674921 ARB07438.1 KY370043 ATP66727.1 MH687968 AYR18599.1 KY370048 ATP66756.1 KF294370 AID16649.1 KY370051 ATP66773.1 KY370049 ATP66762.1 KF294371 AID16655.1 MH687970 AYR18616.1 KF169920 AGO98871.1 AB775894 BAN18703.1 AB671299 BAM15656.1 LC061273 BAS18856.1 LC061274 BAS18866.1 MG757144 AVI15053.1 EU401989 ACB30203.1 MF416379 AWB14621.1 EF446615 ABP87990.1 U14646 AAA87063.1 KF169932 AGO98883.1 AB555560 LC061272 BAJ52885.1 BAS18846.1 KF169923 AGO98874.1 KF169931 AGO98882.1 AY316300 AAQ67205.1 KT318119 KT318120 AMQ23495.1 AMQ23496.1 FJ938067 ACT11030.1 FJ938063 ACT10983.1 GU593319 ADI59790.1 KF169924 AGO98875.1 MN514970 QEY10672.1 JQ173883 AFD97607.1 MN514971 QEY10673.1 KF169930 AGO98881.1 KF169939 KF169940 AGO98890.1 AGO98891.1 FJ415324 ACJ35486.1 JN874559 AFE48795.1 FJ425184 ACJ66946.1 EU814647 ACF21935.1 DQ811784 ABG89288.1 KF169937 AGO98888.1 MH197037 MH197038 MH203065 AXP11688.1 AXP11689.1 AXP11697.1 JN874561 AFE48817.1 KF169915 AGO98866.1 DQ389658 ABD18934.1 KF169933 AGO98884.1 MK046003 QBG67072.1 KF169927 KF169928 KF169929 AGO98878.1 AGO98879.1 AGO98880.1 AF058942 FJ425189 ACJ67000.1 FJ425186 ACJ66971.1 AB593383 BAK18575.1 FJ425185 ACJ66961.1 FJ425190 ACJ67012.1 KF169935 AGO98886.1 KT318116 AMQ23492.1 KT318113 AMQ23489.1 HM573327 HM573328 HM573329 HM573330 ADP21336.1 ADP21337.1 ADP21338.1 ADP21339.1 EF424621 ABP38306.1 KF169938 AGO98889.1 EF445634 ABP73656.1 AY935637 AAX38489.1 KT318124 AMQ23500.1 AF190406 AAF05704.1 FJ938066 ACT11019.1 KF169934 AGO98885.1 MH197039 AXP11690.1 MK046010 QBG67079.1 MH043952 AVZ61099.1 KT318111 AMQ23487.1 EU401986 ACB30200.1 AY935638 AAX38490.1 KT318121 AMQ23497.1
DQ415914 DQ437613 DQ437614 ABD75497.1 ABD75561.1 ABD75569.1 ABD75577.1 ABD75585.1 ABD75641.1 ABD96192.1 ABD96193.1 DQ415900 DQ415901 DQ437618 DQ437619 KF430198 ABD75529.1 ABD75537.1 ABD96197.1 ABD96198.1 AGT17763.1 KF430203 AGT17777.1 DQ415903 ABD75553.1 DQ415908 DQ437607 ABD75593.1 ABD96186.1 DQ415909 DQ437608 ABD75601.1 ABD96187.1 LC315650 BBA20983.1 KF686340 KF686341 KF686342 KF686346 KY674941 KY674942 KY674943 AGW27836.1 AGW27845.1 AGW27854.1 AGW27881.1 ARB07599.1 ARB07608.1 ARB07617.1 KT779555 AMN88686.1 KF686344 AGW27872.1 DQ415910 DQ437609 ABD75609.1 ABD96188.1 KF686343 AGW27863.1 KT779556 AMN88694.1 HM034837 ADN03339.1 KY983584 ARU07577.1 KF430201 AGT17769.1 MK167038 AZS52618.1 DQ339101 DQ415898 DQ437616 ABD75513.1 ABD96195.1 DQ415897 DQ415899 DQ415913 DQ437612 DQ437615 DQ437617 ABD75505.1 ABD75521.1 ABD75633.1 ABD96191.1 ABD96194.1 ABD96196.1 DQ415912 DQ437611 ABD75625.1 ABD96190.1 DQ415902 ABD75545.1 MH940245 AYN64561.1 AY884001 DQ415911 DQ437610 ABD75617.1 ABD96189.1 LC315651 BBA20986.1 KF430197 AGT17758.1 KY674921 ARB07438.1 KY370043 ATP66727.1 MH687968 AYR18599.1 KY370048 ATP66756.1 KF294370 AID16649.1 KY370051 ATP66773.1 KY370049 ATP66762.1 KF294371 AID16655.1 MH687970 AYR18616.1 KF169920 AGO98871.1 AB775894 BAN18703.1 AB671299 BAM15656.1 LC061273 BAS18856.1 LC061274 BAS18866.1 MG757144 AVI15053.1 EU401989 ACB30203.1 MF416379 AWB14621.1 EF446615 ABP87990.1 U14646 AAA87063.1 KF169932 AGO98883.1 AB555560 LC061272 BAJ52885.1 BAS18846.1 KF169923 AGO98874.1 KF169931 AGO98882.1 AY316300 AAQ67205.1 KT318119 KT318120 AMQ23495.1 AMQ23496.1 FJ938067 ACT11030.1 FJ938063 ACT10983.1 GU593319 ADI59790.1 KF169924 AGO98875.1 MN514970 QEY10672.1 JQ173883 AFD97607.1 MN514971 QEY10673.1 KF169930 AGO98881.1 KF169939 KF169940 AGO98890.1 AGO98891.1 FJ415324 ACJ35486.1 JN874559 AFE48795.1 FJ425184 ACJ66946.1 EU814647 ACF21935.1 DQ811784 ABG89288.1 KF169937 AGO98888.1 MH197037 MH197038 MH203065 AXP11688.1 AXP11689.1 AXP11697.1 JN874561 AFE48817.1 KF169915 AGO98866.1 DQ389658 ABD18934.1 KF169933 AGO98884.1 MK046003 QBG67072.1 KF169927 KF169928 KF169929 AGO98878.1 AGO98879.1 AGO98880.1 AF058942 FJ425189 ACJ67000.1 FJ425186 ACJ66971.1 AB593383 BAK18575.1 FJ425185 ACJ66961.1 FJ425190 ACJ67012.1 KF169935 AGO98886.1 KT318116 AMQ23492.1 KT318113 AMQ23489.1 HM573327 HM573328 HM573329 HM573330 ADP21336.1 ADP21337.1 ADP21338.1 ADP21339.1 EF424621 ABP38306.1 KF169938 AGO98889.1 EF445634 ABP73656.1 AY935637 AAX38489.1 KT318124 AMQ23500.1 AF190406 AAF05704.1 FJ938066 ACT11019.1 KF169934 AGO98885.1 MH197039 AXP11690.1 MK046010 QBG67079.1 MH043952 AVZ61099.1 KT318111 AMQ23487.1 EU401986 ACB30200.1 AY935638 AAX38490.1 KT318121 AMQ23497.1
Proteomes
UP000008170
UP000102058
UP000103177
UP000107950
UP000118444
UP000123677
+ More
UP000161305 UP000120443 UP000147573 UP000148743 UP000164754 UP000174991 UP000098607 UP000143896 UP000173725 UP000173897 UP000122230 UP000127413 UP000168247 UP000139452 UP000134208 UP000108760 UP000098016 UP000001985 UP000107997 UP000143869 UP000156925 UP000168964 UP000134743 UP000124867 UP000006551 UP000162514 UP000130220 UP000155347 UP000103372 UP000166748 UP000164707 UP000172225 UP000117254 UP000118088 UP000158296 UP000107676 UP000174569 UP000147981 UP000125269 UP000133443 UP000140344 UP000104294 UP000102696 UP000136116 UP000162935
UP000161305 UP000120443 UP000147573 UP000148743 UP000164754 UP000174991 UP000098607 UP000143896 UP000173725 UP000173897 UP000122230 UP000127413 UP000168247 UP000139452 UP000134208 UP000108760 UP000098016 UP000001985 UP000107997 UP000143869 UP000156925 UP000168964 UP000134743 UP000124867 UP000006551 UP000162514 UP000130220 UP000155347 UP000103372 UP000166748 UP000164707 UP000172225 UP000117254 UP000118088 UP000158296 UP000107676 UP000174569 UP000147981 UP000125269 UP000133443 UP000140344 UP000104294 UP000102696 UP000136116 UP000162935
Interpro
SUPFAM
SSF143587
SSF143587
Gene 3D
ProteinModelPortal
Q5MQD0
Q19U42
Q19U37
S5YGG8
Q0ZJH3
Q19U49
+ More
Q19U48 A0A224AU27 U3NAI2 A0A140H1H1 U3N9S7 Q19U47 U3N885 A0A140H1H9 E0YJ44 A0A1Y0EUZ8 S5YA28 A0A451ERM8 Q0ZME7 Q19U40 Q19U39 Q19U45 Q0ZJI1 A0A3G2KXL1 Q14EB0 Q19U46 A0A224ANC9 S5ZBP3 A0A1V0E0Z4 A0A2H4MWY0 A0A3G3NGZ3 A0A2H4MX03 A0A096XNG8 A0A2H4MZ72 A0A2H4MZ10 A0A096XNI4 A0A3G3NHC8 S4WG83 R4WAW7 I2FKH1 A0A0K2RVK5 A0A0K2RVL1 A0A3S7GYI2 B9TXV2 A0A2S0SZ25 A8R4D0 Q83331 S4WIX3 E5RPZ2 S4WLK1 S4WML5 Q6W355 A0A142D826 C6GHR0 C6GHL3 D7PJY2 S4WHI9 A0A5J6NUL2 H9BZX9 A0A5J6NRR8 S4WG91 S4WG99 B7TYG9 H9AA34 B7U2L1 B3VTS5 Q0PL39 S4WIX5 A0A346I7G9 H9AA55 S4WG79 Q2EEY3 S4WLK8 A0A481S1P4 S4WLK4 P25192 B7U2R5 B7U2N3 F1T0N1 B7U2M2 B7U2S7 S4WG94 A0A142D823 A0A142D820 E3UTE9 A4ZU41 S4WLL0 A6YD40 Q58NS2 A0A142D831 Q9QD01 C6GHP9 S4WHJ6 A0A346I7H0 A0A481S1B7 A0A2R4STR1 A0A142D818 B9TXU9 Q58NS1 A0A142D828
Q19U48 A0A224AU27 U3NAI2 A0A140H1H1 U3N9S7 Q19U47 U3N885 A0A140H1H9 E0YJ44 A0A1Y0EUZ8 S5YA28 A0A451ERM8 Q0ZME7 Q19U40 Q19U39 Q19U45 Q0ZJI1 A0A3G2KXL1 Q14EB0 Q19U46 A0A224ANC9 S5ZBP3 A0A1V0E0Z4 A0A2H4MWY0 A0A3G3NGZ3 A0A2H4MX03 A0A096XNG8 A0A2H4MZ72 A0A2H4MZ10 A0A096XNI4 A0A3G3NHC8 S4WG83 R4WAW7 I2FKH1 A0A0K2RVK5 A0A0K2RVL1 A0A3S7GYI2 B9TXV2 A0A2S0SZ25 A8R4D0 Q83331 S4WIX3 E5RPZ2 S4WLK1 S4WML5 Q6W355 A0A142D826 C6GHR0 C6GHL3 D7PJY2 S4WHI9 A0A5J6NUL2 H9BZX9 A0A5J6NRR8 S4WG91 S4WG99 B7TYG9 H9AA34 B7U2L1 B3VTS5 Q0PL39 S4WIX5 A0A346I7G9 H9AA55 S4WG79 Q2EEY3 S4WLK8 A0A481S1P4 S4WLK4 P25192 B7U2R5 B7U2N3 F1T0N1 B7U2M2 B7U2S7 S4WG94 A0A142D823 A0A142D820 E3UTE9 A4ZU41 S4WLL0 A6YD40 Q58NS2 A0A142D831 Q9QD01 C6GHP9 S4WHJ6 A0A346I7H0 A0A481S1B7 A0A2R4STR1 A0A142D818 B9TXU9 Q58NS1 A0A142D828
PDB
5I08
E-value=0,
Score=5278
Ontologies
KEGG
GO
GO:0019031 C:viral envelope
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0016021 C:integral component of membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0020002 C:host cell plasma membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0042802 F:identical protein binding
GO:0061025 P:membrane fusion
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0016021 C:integral component of membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0020002 C:host cell plasma membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0042802 F:identical protein binding
GO:0061025 P:membrane fusion
Subcellular Location
From MSLVP
Host nucleus > host nucleolus. Host cytoplasm. Secreted.
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
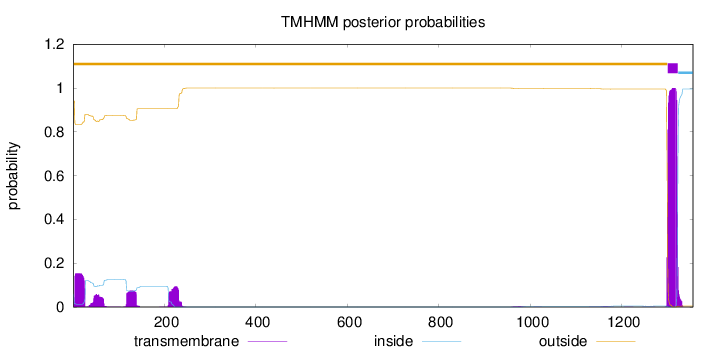
Topology
Length:
1356
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
31.6869199999999
Exp number, first 60 AAs:
4.43846
Total prob of N-in:
0.05951
outside
1 - 1300
TMhelix
1301 - 1323
inside
1324 - 1356
Population Genetic Test Statistics
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 |
Protein |
YP_173238.1 |
|
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.