Strain
CHINA_BAT_2006_NC_009020
(Region: China: Guangdong province; Strain: Bat coronavirus HKU5-1, complete genome.; Date: 2006)
Gene
spike glycoprotein
Description
Annotated in NCBI,
spike glycoprotein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGATACGCTCAGTGTTAGTACTGATGTGCTCGTTAACTTTTATAGGAAACCTCACAAGAGGTCAAAGTGTTGATATGGGACACAATGGCACTGGTTCATGTTTAGATTCCCAGGTACAACCTGATTATTTTGAATCTGTGCATACTACTTGGCCCATGCCTATTGACACGAGTAAGGCTGAAGGTGTCATTTATCCTAATGGCAAGTCCTACTCTAATATTACTCTAACTTATACGGGACTGTACCCCAAGGCTAATGACCTTGGTAAACAGTATTTATTCTCTGACGGACATAGTGCCCCAGGACGTCTTAATAACTTATTTGTCAGTAATTACTCCTCACAAGTGGAGTCCTTTGATGATGGCTTTGTGGTCCGTATTGGTGCTGCTGCCAATAAAACTGGAACCACTGTCATATCTCAATCCACATTTAAACCCATTAAGAAAATCTATCCAGCCTTTTTGCTTGGACATTCTGTGGGTAATTATACACCGTCCAATAGGACAGGTCGTTATCTCAACCACACGCTTGTCATCCTACCGGATGGCTGTGGTACTATTTTACATGCATTCTATTGTGTTCTTCACCCTCGCACACAACAAAATTGTGCTGGTGAAACAAATTTCAAATCCCTCTCGCTTTGGGATACTCCTGCATCAGACTGTGTCTCTGGTTCTTATAACCAAGAAGCCACATTAGGTGCTTTCAAAGTGTATTTTGACTTGATTAATTGCACGTTCAGATACAATTATACTATAACAGAGGATGAAAACGCTGAGTGGTTCGGCATTACCCAAGACACACAAGGCGTTCACCTCTATTCATCTCGAAAAGAGAACGTGTTCAGAAATAACATGTTCCACTTTGCTACTTTACCTGTGTACCAGAAAATCCTCTATTACACAGTCATTCCTCGCAGCATCCGAAGTCCTTTCAATGACAGAAAAGCCTGGGCTGCATTCTACATTTATAAACTACATCCACTCACATATTTGCTAAATTTTGATGTGGAGGGCTATATAACTAAAGCTGTAGACTGTGGCTACGATGATCTAGCACAGCTACAATGTTCCTACGAATCCTTCGAAGTCGAAACAGGCGTTTATTCCGTTTCATCGTTCGAAGCTTCTCCTAGAGGTGAGTTCATTGAACAGGCAACAACTCAAGAGTGTGATTTCACACCTATGTTGACTGGTACACCTCCTCCTATATATAATTTCAAAAGGTTGGTCTTCACCAATTGCAATTACAACTTAACAAAACTCCTTTCACTGTTTCAAGTGAGCGAGTTTTCTTGTCATCAAGTTTCACCTAGTAGTCTTGCTACCGGTTGTTACTCTTCTCTTACAGTGGACTATTTCGCTTATTCCACTGATATGAGTTCTTACCTGCAACCTGGCTCCGCTGGAGCAATTGTGCAGTTTAATTACAAACAAGACTTTAGCAATCCCACGTGTAGAGTGCTTGCTACTGTTCCACAAAATCTTACTACAATTACTAAACCTAGTAATTATGCTTATCTTACAGAGTGTTATAAAACCAGTGCATATGGCAAGAATTACTTGTACAATGCGCCTGGCGCTTACACTCCTTGCTTATCTTTAGCCTCTCGTGGGTTCTCTACTAAATACCAGTCACATAGTGATGGCGAGCTAACCACTACTGGTTACATTTATCCTGTCACTGGAAATCTTCAAATGGCTTTCATAATTTCTGTTCAGTATGGAACTGACACTAACAGTGTCTGCCCCATGCAAGCATTAAGAAATGATACTAGCATTGAAGATAAGCTAGACGTTTGTGTTGAATACTCGCTCCATGGTATAACTGGAAGGGGGGTTTTCCACAATTGCACATCTGTTGGACTGAGAAACCAGCGGTTTGTGTATGATACTTTCGACAATTTAGTTGGTTACCATTCTGACAATGGTAATTACTATTGTGTCAGACCTTGTGTCAGTGTGCCTGTCTCTGTGATTTACGACAAGGCATCTAATTCTCATGCTACATTATTCGGAAGCGTTGCATGTTCGCACGTTACCACAATGATGTCGCAATTTTCACGCATGACCAAAACTAATTTGCTCGCGCGTACAACTCCAGGTCCATTGCAAACTACTGTTGGTTGTGCAATGGGCTTTATCAATTCCTCAATGGTAGTTGACGAATGTCAACTTCCGCTTGGTCAATCACTTTGTGCTATTCCACCAACTACTTCTTCACGCGTTCGACGTGCTACTTCTGGTGCATCTGATGTGTTTCAAATCGCCACTCTTAACTTTACTAGTCCATTAACACTCGCACCAATAAATTCTACTGGATTTGTTGTTGCTGTGCCGACTAATTTCACATTTGGTGTCACTCAAGAATTCATTGAGACTACCATTCAAAAGATTACTGTCGATTGCAAGCAGTACGTTTGTAATGGTTTTAAGAAGTGCGAAGACTTGCTCAAAGAATATGGCCAGTTTTGCTCTAAAATTAACCAGGCTCTTCATGGTGCAAACCTACGTCAAGACGAGTCTATTGCTAATCTATTTTCAAGTATTAAAACCCAGAATACCCAGCCTCTTCAGGCGGGATTGAATGGTGATTTTAACTTGACTATGCTTCAAATACCTCAAGTTACTACAGGTGAACGTAAGTACAGGAGTACTATCGAAGACCTTCTTTTCAATAAGGTTACTATTGCTGATCCTGGCTATATGCAAGGCTATGATGAATGTATGCAGCAAGGTCCTCAGTCAGCTCGAGACTTGATCTGTGCACAATATGTTGCTGGCTACAAAGTGCTGCCACCCTTATATGACCCCTATATGGAAGCTGCTTACACTTCTTCCCTATTGGGCAGTATTGCTGGTGCTAGTTGGACAGCAGGTCTGTCGTCTTTCGCAGCAATACCATTTGCACAAAGTATCTTTTATCGCTTGAATGGTGTTGGCATCACTCAGCAGGTTCTGTCTGAGAATCAGAAGATCATTGCCAACAAATTCAATCAAGCTCTTGGTGCCATGCAAACTGGCTTTACTACTACTAACCTTGCTTTCAACAAGGTTCAGGATGCAGTAAATGCTAATGCAATGGCTCTTTCCAAACTAGCTGCGGAATTGTCTAACACTTTCGGTGCTATTTCATCATCAATCAGTGACATCCTTGCAAGGCTTGACACTGTTGAACAAGAGGCTCAAATTGATCGGTTGATCAATGGACGTCTTACATCCCTTAACGCATTCGTTGCGCAACAACTCGTGCGTACTGAAGCTGCTGCTAGATCTGCCCAATTGGCTCAAGATAAGGTCAATGAGTGCGTGAAGTCGCAATCCAAACGGAATGGATTCTGCGGAACTGGCACACATATTGTTTCATTTGCCATTAATGCTCCTAATGGCCTCTACTTCTTCCATGTTGGTTACCAGCCAACATCCCATGTCAATGCAACTGCCGCTTATGGCCTTTGCAACACTGAAAATCCACAAAAGTGCATTGCACCTATTGATGGATACTTCGTCTTAAACCAAACTACCAGCACTGTCGCAGACAGTGACCAACAATGGTATTATACTGGTAGTTCCTTCTTCCATCCTGAACCCATCACAGAAGCAAATTCTAAGTATGTGTCCATGGATGTAAAATTCGAAAACCTCACTAATAGGCTTCCTCCACCGCTCCTTAGTAATTCAACAGATTTGGATTTCAAGGAAGAGCTGGAAGAATTCTTCAAAAACGTCTCCTCACAAGGACCAAACTTCCAGGAGATTTCTAAGATTAACACTACACTGCTTAACCTCAACACAGAGTTGATGGTGCTGAGTGAAGTTGTTAAACAATTAAATGAATCCTATATCGACTTGAAAGAGTTGGGGAATTATACTTTTTACCAAAAATGGCCATGGTATATATGGCTTGGCTTTATTGCAGGGCTTGTTGCTCTTGCTCTTTGTGTGTTCTTCATCTTATGCTGTACTGGCTGTGGTACTAGCTGTTTGGGGAAACTAAAATGTAATCGTTGTTGTGACTCGTATGATGAGTATGAGGTCGAGAAGATCCATGTTCATTGA
Protein
MIRSVLVLMCSLTFIGNLTRGQSVDMGHNGTGSCLDSQVQPDYFESVHTTWPMPIDTSKAEGVIYPNGKSYSNITLTYTGLYPKANDLGKQYLFSDGHSAPGRLNNLFVSNYSSQVESFDDGFVVRIGAAANKTGTTVISQSTFKPIKKIYPAFLLGHSVGNYTPSNRTGRYLNHTLVILPDGCGTILHAFYCVLHPRTQQNCAGETNFKSLSLWDTPASDCVSGSYNQEATLGAFKVYFDLINCTFRYNYTITEDENAEWFGITQDTQGVHLYSSRKENVFRNNMFHFATLPVYQKILYYTVIPRSIRSPFNDRKAWAAFYIYKLHPLTYLLNFDVEGYITKAVDCGYDDLAQLQCSYESFEVETGVYSVSSFEASPRGEFIEQATTQECDFTPMLTGTPPPIYNFKRLVFTNCNYNLTKLLSLFQVSEFSCHQVSPSSLATGCYSSLTVDYFAYSTDMSSYLQPGSAGAIVQFNYKQDFSNPTCRVLATVPQNLTTITKPSNYAYLTECYKTSAYGKNYLYNAPGAYTPCLSLASRGFSTKYQSHSDGELTTTGYIYPVTGNLQMAFIISVQYGTDTNSVCPMQALRNDTSIEDKLDVCVEYSLHGITGRGVFHNCTSVGLRNQRFVYDTFDNLVGYHSDNGNYYCVRPCVSVPVSVIYDKASNSHATLFGSVACSHVTTMMSQFSRMTKTNLLARTTPGPLQTTVGCAMGFINSSMVVDECQLPLGQSLCAIPPTTSSRVRRATSGASDVFQIATLNFTSPLTLAPINSTGFVVAVPTNFTFGVTQEFIETTIQKITVDCKQYVCNGFKKCEDLLKEYGQFCSKINQALHGANLRQDESIANLFSSIKTQNTQPLQAGLNGDFNLTMLQIPQVTTGERKYRSTIEDLLFNKVTIADPGYMQGYDECMQQGPQSARDLICAQYVAGYKVLPPLYDPYMEAAYTSSLLGSIAGASWTAGLSSFAAIPFAQSIFYRLNGVGITQQVLSENQKIIANKFNQALGAMQTGFTTTNLAFNKVQDAVNANAMALSKLAAELSNTFGAISSSISDILARLDTVEQEAQIDRLINGRLTSLNAFVAQQLVRTEAAARSAQLAQDKVNECVKSQSKRNGFCGTGTHIVSFAINAPNGLYFFHVGYQPTSHVNATAAYGLCNTENPQKCIAPIDGYFVLNQTTSTVADSDQQWYYTGSSFFHPEPITEANSKYVSMDVKFENLTNRLPPPLLSNSTDLDFKEELEEFFKNVSSQGPNFQEISKINTTLLNLNTELMVLSEVVKQLNESYIDLKELGNYTFYQKWPWYIWLGFIAGLVALALCVFFILCCTGCGTSCLGKLKCNRCCDSYDEYEVEKIHVH
Summary
Function
attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S1: attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S1: attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes.
Similarity
Belongs to the betacoronaviruses spike protein family.
Keywords
3D-structure
Coiled coil
Disulfide bond
Fusion of virus membrane with host endosomal membrane
Fusion of virus membrane with host membrane
Glycoprotein
Host cell membrane
Host membrane
Host-virus interaction
Lipoprotein
Membrane
Palmitate
Reference proteome
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Viral penetration into host cytoplasm
Virion
Virulence
Virus entry into host cell
Feature
chain Spike glycoprotein
Uniprot
A3EXD0
S4WZQ4
S4X422
S4WWR8
S4WZR3
S4WWS3
+ More
A0A0U1WHM0 S4WZR0 S4X278 S4X284 S4WWR5 A3EXD9 A3EXE8 A3EXF7 S4WYF2 S4WYE5 S4X419 S4WYE8 S4X425 S4X276 Q0Q4F7 A0A2I6PIW5 A0A2I6PIX8 A0A023Y9K3 A0A2I4R8V9 A0A2I4R888 A0A2R4KP93 A0A2R4KP86 A0A5H2WTJ3 A0A1Z1W230 A0A1Z1W244 S4WYD2 Q0Q4F2 A0A1Z1W227 S4WWQ3 S4WZQ1 A0A166ZLN4 S4WYD6 A3EXA3 A3EX94 A3EXC1 A3EXB2 A0A0U1WJZ6 A0A140AZ05 A0A0A0Q4U2 A0A0P0HTL4 A0A291I6G6 A0A0U2J5T6 A0A0U2INY2 A0A126G7Z3 A0A0U2MGJ8 A0A0U2MS80 A0A2H1YPP7 A0A0P0GPV1 A0A140AQ46 A0A0U2NTX5 A0A0U4AGH8 A0A0P0HJE7 A0A2D0YM48 A0A2D0YGK8 A0A0P0HBE6 A0A0U2P9C9 A0A0A0Q7F8 A0A0U2NE92 A0A0A0Q7G4 A0A2D0YIQ0 A0A1D6Y6X4 A0A0U2P188 A0A0U2J5U2 A0A2D0YCT4 T2B9T0 A0A0U2GMX5 T2B999 A0A0U2GLS2 W5ZZF5 A0A0U2MS90 A0A0U2NXW5 A0A3T0I8D8 A0A0G3U597 A0A2D0YCE0 R9UCW7 A0A1C8EAZ2 A0A0U2GPS7 A0A0U2P195 A0A291I6B7 A0A3S7R2I2 K0BRG7 A0A384VXU1 W6A028 A0A411PBL8 A0A2D0YNJ0 A0A2D0YPZ8 A0A2D0YAP4 A0A0U3SQE0 A0A2D0Y380 A0A0U2NE84 A0A2D0YDA3 A0A1W5LGU2 A0A5J6ED89 A0A140AZ15
A0A0U1WHM0 S4WZR0 S4X278 S4X284 S4WWR5 A3EXD9 A3EXE8 A3EXF7 S4WYF2 S4WYE5 S4X419 S4WYE8 S4X425 S4X276 Q0Q4F7 A0A2I6PIW5 A0A2I6PIX8 A0A023Y9K3 A0A2I4R8V9 A0A2I4R888 A0A2R4KP93 A0A2R4KP86 A0A5H2WTJ3 A0A1Z1W230 A0A1Z1W244 S4WYD2 Q0Q4F2 A0A1Z1W227 S4WWQ3 S4WZQ1 A0A166ZLN4 S4WYD6 A3EXA3 A3EX94 A3EXC1 A3EXB2 A0A0U1WJZ6 A0A140AZ05 A0A0A0Q4U2 A0A0P0HTL4 A0A291I6G6 A0A0U2J5T6 A0A0U2INY2 A0A126G7Z3 A0A0U2MGJ8 A0A0U2MS80 A0A2H1YPP7 A0A0P0GPV1 A0A140AQ46 A0A0U2NTX5 A0A0U4AGH8 A0A0P0HJE7 A0A2D0YM48 A0A2D0YGK8 A0A0P0HBE6 A0A0U2P9C9 A0A0A0Q7F8 A0A0U2NE92 A0A0A0Q7G4 A0A2D0YIQ0 A0A1D6Y6X4 A0A0U2P188 A0A0U2J5U2 A0A2D0YCT4 T2B9T0 A0A0U2GMX5 T2B999 A0A0U2GLS2 W5ZZF5 A0A0U2MS90 A0A0U2NXW5 A0A3T0I8D8 A0A0G3U597 A0A2D0YCE0 R9UCW7 A0A1C8EAZ2 A0A0U2GPS7 A0A0U2P195 A0A291I6B7 A0A3S7R2I2 K0BRG7 A0A384VXU1 W6A028 A0A411PBL8 A0A2D0YNJ0 A0A2D0YPZ8 A0A2D0YAP4 A0A0U3SQE0 A0A2D0Y380 A0A0U2NE84 A0A2D0YDA3 A0A1W5LGU2 A0A5J6ED89 A0A140AZ15
Pubmed
EMBL
EF065509
KC522090
AGP04929.1
KC522099
AGP04938.1
KC522098
+ More
AGP04937.1 KC522100 AGP04939.1 KC522103 AGP04942.1 KJ473820 AIA62343.1 KC522095 AGP04934.1 KC522096 AGP04935.1 KC522101 AGP04940.1 KC522093 AGP04932.1 EF065510 ABN10884.1 EF065511 ABN10893.1 EF065512 ABN10902.1 KC522102 AGP04941.1 KC522092 AGP04931.1 KC522094 AGP04933.1 KC522097 AGP04936.1 KC522104 AGP04943.1 KC522091 AGP04930.1 DQ648790 ABG11962.1 MG596802 AUM60014.1 MG596803 AUM60024.1 KJ473821 AHY61337.1 KX442564 ASL68941.1 KX442565 ASL68953.1 MG021452 AVV62537.1 MG021451 AVV62526.1 LC469308 BBJ36008.1 KY783843 ARX80139.1 KY783844 ARX80140.1 KC522049 AGP04916.1 DQ648794 KY783842 ARX80138.1 KC522050 AGP04917.1 KC522057 AGP04924.1 KU182965 ANA96039.1 KC522051 KC522052 KC522053 KC522054 KC522055 KC522056 KC522058 KC522059 KC522060 KC522061 AGP04918.1 AGP04919.1 AGP04920.1 AGP04921.1 AGP04922.1 AGP04923.1 AGP04925.1 AGP04926.1 AGP04927.1 AGP04928.1 EF065506 ABN10848.1 EF065505 EF065508 ABN10866.1 EF065507 ABN10857.1 KJ473822 AIA62352.1 KT868875 KX034094 KX034095 ALK80291.1 ANC28645.1 ANC28656.1 KM027279 KT368824 AID55090.1 ALA49341.1 KR912188 KR912195 KU710264 ALJ76278.1 AMQ48993.1 MG011356 ATG84855.1 KT806006 ALJ54486.1 KT806041 ALJ54521.1 KT374057 ALB08322.1 KT806016 ALJ54496.1 KT805987 ALJ54467.1 MF598597 ASU89955.1 KR912187 ALJ76277.1 KT374056 ALB08311.1 KT806028 ALJ54508.1 KT806049 ALW82691.1 KR912196 ALJ76286.1 MF598638 ASU90406.1 MF598608 MF598650 MF598700 ASU90076.1 ASU90538.1 ASU91087.1 KT368828 KT368843 KT368844 KT368845 KT368849 KT368850 KT368851 KT368855 KT368887 KT368888 KT805995 KT805997 KT805998 KT805999 KT806000 KT806013 KR912189 KR912190 KR912191 KR912192 KR912193 KR912194 KT877351 ALA49385.1 ALA49550.1 ALA49561.1 ALA49572.1 ALA49616.1 ALA49627.1 ALA49638.1 ALA49682.1 ALA50034.1 ALA50045.1 ALJ54475.1 ALJ54477.1 ALJ54478.1 ALJ54479.1 ALJ54480.1 ALJ54493.1 ALJ76282.1 ALL26409.1 KT805970 KT806001 KT806004 ALJ54450.1 ALJ54481.1 ALJ54484.1 KM027273 AID55084.1 KT806010 ALJ54490.1 KM027288 AID55099.1 MF598711 ASU91208.1 KT861627 ALX27232.1 KT806021 ALJ54501.1 KT806038 ALJ54518.1 MF598641 ASU90439.1 KF600652 KT806040 AGV08584.1 ALJ54520.1 KT368880 KT368881 ALA49957.1 ALA49968.1 KF600612 KF600620 AGV08379.1 AGV08408.1 KT368866 KT368867 ALA49803.1 ALA49814.1 KF917527 KF958702 KJ156949 KJ156952 KJ556336 AHE78097.1 AHE78108.1 AHI48594.1 AHI48605.1 AHN10812.1 KT805978 KT806003 ALJ54458.1 ALJ54483.1 KT805972 KT806055 ALJ54452.1 ALW82753.1 MK280984 AZU90731.1 KT029139 AKL59401.1 MF598631 ASU90329.1 KF192507 AGN52936.1 KT877350 ALL26396.1 KT368852 ALA49649.1 KT806037 ALJ54517.1 MG011354 ATG84833.1 MG757597 AXN73437.1 JX869059 AFS88936.1 MF741827 MF741832 ASY99811.1 ASY99864.1 KJ156934 KJ156873 KJ713295 KJ713296 KM015348 KM027281 KM210278 KP966104 KT006149 KT026453 KT026454 KT026455 KT026456 KT036372 KT036373 KT036374 KT182953 KT275306 KT275307 KT275308 KT368833 KT368868 KT368870 KT368871 KT368872 KT368873 KT368874 KT368876 KT368877 KT368878 KT368882 KT368883 KT368884 KT368885 KT368886 KT368889 KT805964 KT805965 KT805967 KT805969 KT805974 KT805977 KT805979 KT805980 KT805982 KT805984 KT805989 KT805990 KT805993 KT805996 KT806002 KT806005 KT806007 KT806009 KT806012 KT806014 KT806018 KT806019 KT806023 KT806024 KT806025 KT806026 KT806027 KT806029 KT806030 KT806031 KT806033 KT806034 KT806035 KT806039 KT868870 KU233362 KU233363 KU233364 KT806044 KT806045 KT806046 KT806047 KT806048 KT806050 KT806051 KT806052 KT806053 KU851859 KU851860 KU851861 KU851862 KU851863 KU851864 KX154684 KX108945 KY688118 KY688120 KY688121 KY688122 KY688123 KY688124 MF598594 MF598595 MF598603 MF598604 MF598606 MF598607 MF598611 MF598612 MF598613 MF598615 MF598620 MF598625 MF598627 MF598628 MF598633 MF598643 MF598644 MF598645 MF598646 MF598652 MF598654 MF598657 MF598660 MF598661 MF598662 MF598664 MF598665 MF598666 MF598668 MF598670 MF598677 MF598680 MF598682 MF598684 MF598685 MF598686 MF598687 MF598688 MF598691 MF598692 MF598694 MF598695 MF598697 MF598698 MF598701 MF741837 MF741825 MF741826 MF741828 MF741829 MF741831 MF741833 MF741834 MF741835 MF741836 MG011343 MG011352 MG011353 MG011355 MF679172 MG366880 MG912608 MH306207 MH371127 MH432120 MG757593 MG757594 MG757595 MG757596 MG757598 MG757599 MG757600 MG757601 MG757602 MG757603 MH029552 MK129253 MH978888 MK462248 MH259486 MK858156 MK858158 MK858163 AHI48572.1 AHI48662.1 AHY22525.1 AHY22535.1 AID50418.1 AID55092.1 AJD81451.1 AKA63509.1 AKJ80137.2 AKK52582.1 AKK52592.1 AKK52602.1 AKK52612.1 AKL80593.1 AKL80604.1 AKL80615.1 AKN11071.1 AKQ21055.1 AKQ21064.1 AKQ21073.1 ALA49440.1 ALA49825.1 ALA49847.1 ALA49858.1 ALA49869.1 ALA49880.1 ALA49891.1 ALA49913.1 ALA49924.1 ALA49935.1 ALA49979.1 ALA49990.1 ALA50001.1 ALA50012.1 ALA50023.1 ALA50056.1 ALJ54444.1 ALJ54445.1 ALJ54447.1 ALJ54449.1 ALJ54454.1 ALJ54457.1 ALJ54459.1 ALJ54460.1 ALJ54462.1 ALJ54464.1 ALJ54469.1 ALJ54470.1 ALJ54473.1 ALJ54476.1 ALJ54482.1 ALJ54485.1 ALJ54487.1 ALJ54489.1 ALJ54492.1 ALJ54494.1 ALJ54498.1 ALJ54499.1 ALJ54503.1 ALJ54504.1 ALJ54505.1 ALJ54506.1 ALJ54507.1 ALJ54509.1 ALJ54510.1 ALJ54511.1 ALJ54513.1 ALJ54514.1 ALJ54515.1 ALJ54519.1 ALK80242.1 ALT66802.1 ALT66813.1 ALT66824.1 ALW82636.1 ALW82647.1 ALW82658.1 ALW82669.1 ALW82680.1 ALW82702.1 ALW82709.1 ALW82720.1 ALW82731.1 AMQ49015.1 AMQ49026.1 AMQ49037.1 AMQ49048.1 AMQ49059.1 AMQ49070.1 ANF29162.1 ANI69911.1 AQZ41311.1 AQZ41332.1 AQZ41343.1 AQZ41354.1 AQZ41365.1 AQZ41376.1 ASU89921.1 ASU89933.1 ASU90021.1 ASU90032.1 ASU90054.1 ASU90065.1 ASU90109.1 ASU90120.1 ASU90131.1 ASU90153.1 ASU90208.1 ASU90263.1 ASU90285.1 ASU90295.1 ASU90351.1 ASU90461.1 ASU90472.1 ASU90483.1 ASU90494.1 ASU90560.1 ASU90582.1 ASU90615.1 ASU90648.1 ASU90659.1 ASU90670.1 ASU90692.1 ASU90703.1 ASU90714.1 ASU90736.1 ASU90758.1 ASU90835.1 ASU90868.1 ASU90890.1 ASU90912.1 ASU90923.1 ASU90934.1 ASU90945.1 ASU90956.1 ASU90988.1 ASU90999.1 ASU91021.1 ASU91032.1 ASU91054.1 ASU91065.1 ASU91098.1 ASY99778.1 ASY99789.1 ASY99800.1 ASY99820.1 ASY99831.1 ASY99853.1 ASY99873.1 ASY99884.1 ASY99895.1 ASY99906.1 ATG84712.1 ATG84811.1 ATG84822.1 ATG84844.1 ATW75478.1 ATY74381.1 AVK87461.1 AWM99582.1 AXG21643.1 AXG21665.1 AXN73393.1 AXN73404.1 AXN73415.1 AXN73426.1 AXN73448.1 AXN73459.1 AXN73470.1 AXN73481.1 AXN73492.1 AXN73503.1 AYO86699.1 AZK15900.1 QBF44115.1 QBF80510.1 QCI31480.1 QCQ28828.1 QCQ28830.1 QCQ28835.1 MK462253 MK462254 MK462255 QBF80565.1 QBF80576.1 QBF80587.1 MF598600 MF598678 ASU89988.1 ASU90846.1 MF598632 ASU90340.1 MF598679 ASU90857.1 KT806054 ALW82742.1 MF598623 ASU90241.1 KT805994 ALJ54474.1 MF598651 ASU90549.1 KX154693 ANF29261.1 MN403102 QEU56412.1 KT868872 KT868876 ALK80261.1 ALK80301.1
AGP04937.1 KC522100 AGP04939.1 KC522103 AGP04942.1 KJ473820 AIA62343.1 KC522095 AGP04934.1 KC522096 AGP04935.1 KC522101 AGP04940.1 KC522093 AGP04932.1 EF065510 ABN10884.1 EF065511 ABN10893.1 EF065512 ABN10902.1 KC522102 AGP04941.1 KC522092 AGP04931.1 KC522094 AGP04933.1 KC522097 AGP04936.1 KC522104 AGP04943.1 KC522091 AGP04930.1 DQ648790 ABG11962.1 MG596802 AUM60014.1 MG596803 AUM60024.1 KJ473821 AHY61337.1 KX442564 ASL68941.1 KX442565 ASL68953.1 MG021452 AVV62537.1 MG021451 AVV62526.1 LC469308 BBJ36008.1 KY783843 ARX80139.1 KY783844 ARX80140.1 KC522049 AGP04916.1 DQ648794 KY783842 ARX80138.1 KC522050 AGP04917.1 KC522057 AGP04924.1 KU182965 ANA96039.1 KC522051 KC522052 KC522053 KC522054 KC522055 KC522056 KC522058 KC522059 KC522060 KC522061 AGP04918.1 AGP04919.1 AGP04920.1 AGP04921.1 AGP04922.1 AGP04923.1 AGP04925.1 AGP04926.1 AGP04927.1 AGP04928.1 EF065506 ABN10848.1 EF065505 EF065508 ABN10866.1 EF065507 ABN10857.1 KJ473822 AIA62352.1 KT868875 KX034094 KX034095 ALK80291.1 ANC28645.1 ANC28656.1 KM027279 KT368824 AID55090.1 ALA49341.1 KR912188 KR912195 KU710264 ALJ76278.1 AMQ48993.1 MG011356 ATG84855.1 KT806006 ALJ54486.1 KT806041 ALJ54521.1 KT374057 ALB08322.1 KT806016 ALJ54496.1 KT805987 ALJ54467.1 MF598597 ASU89955.1 KR912187 ALJ76277.1 KT374056 ALB08311.1 KT806028 ALJ54508.1 KT806049 ALW82691.1 KR912196 ALJ76286.1 MF598638 ASU90406.1 MF598608 MF598650 MF598700 ASU90076.1 ASU90538.1 ASU91087.1 KT368828 KT368843 KT368844 KT368845 KT368849 KT368850 KT368851 KT368855 KT368887 KT368888 KT805995 KT805997 KT805998 KT805999 KT806000 KT806013 KR912189 KR912190 KR912191 KR912192 KR912193 KR912194 KT877351 ALA49385.1 ALA49550.1 ALA49561.1 ALA49572.1 ALA49616.1 ALA49627.1 ALA49638.1 ALA49682.1 ALA50034.1 ALA50045.1 ALJ54475.1 ALJ54477.1 ALJ54478.1 ALJ54479.1 ALJ54480.1 ALJ54493.1 ALJ76282.1 ALL26409.1 KT805970 KT806001 KT806004 ALJ54450.1 ALJ54481.1 ALJ54484.1 KM027273 AID55084.1 KT806010 ALJ54490.1 KM027288 AID55099.1 MF598711 ASU91208.1 KT861627 ALX27232.1 KT806021 ALJ54501.1 KT806038 ALJ54518.1 MF598641 ASU90439.1 KF600652 KT806040 AGV08584.1 ALJ54520.1 KT368880 KT368881 ALA49957.1 ALA49968.1 KF600612 KF600620 AGV08379.1 AGV08408.1 KT368866 KT368867 ALA49803.1 ALA49814.1 KF917527 KF958702 KJ156949 KJ156952 KJ556336 AHE78097.1 AHE78108.1 AHI48594.1 AHI48605.1 AHN10812.1 KT805978 KT806003 ALJ54458.1 ALJ54483.1 KT805972 KT806055 ALJ54452.1 ALW82753.1 MK280984 AZU90731.1 KT029139 AKL59401.1 MF598631 ASU90329.1 KF192507 AGN52936.1 KT877350 ALL26396.1 KT368852 ALA49649.1 KT806037 ALJ54517.1 MG011354 ATG84833.1 MG757597 AXN73437.1 JX869059 AFS88936.1 MF741827 MF741832 ASY99811.1 ASY99864.1 KJ156934 KJ156873 KJ713295 KJ713296 KM015348 KM027281 KM210278 KP966104 KT006149 KT026453 KT026454 KT026455 KT026456 KT036372 KT036373 KT036374 KT182953 KT275306 KT275307 KT275308 KT368833 KT368868 KT368870 KT368871 KT368872 KT368873 KT368874 KT368876 KT368877 KT368878 KT368882 KT368883 KT368884 KT368885 KT368886 KT368889 KT805964 KT805965 KT805967 KT805969 KT805974 KT805977 KT805979 KT805980 KT805982 KT805984 KT805989 KT805990 KT805993 KT805996 KT806002 KT806005 KT806007 KT806009 KT806012 KT806014 KT806018 KT806019 KT806023 KT806024 KT806025 KT806026 KT806027 KT806029 KT806030 KT806031 KT806033 KT806034 KT806035 KT806039 KT868870 KU233362 KU233363 KU233364 KT806044 KT806045 KT806046 KT806047 KT806048 KT806050 KT806051 KT806052 KT806053 KU851859 KU851860 KU851861 KU851862 KU851863 KU851864 KX154684 KX108945 KY688118 KY688120 KY688121 KY688122 KY688123 KY688124 MF598594 MF598595 MF598603 MF598604 MF598606 MF598607 MF598611 MF598612 MF598613 MF598615 MF598620 MF598625 MF598627 MF598628 MF598633 MF598643 MF598644 MF598645 MF598646 MF598652 MF598654 MF598657 MF598660 MF598661 MF598662 MF598664 MF598665 MF598666 MF598668 MF598670 MF598677 MF598680 MF598682 MF598684 MF598685 MF598686 MF598687 MF598688 MF598691 MF598692 MF598694 MF598695 MF598697 MF598698 MF598701 MF741837 MF741825 MF741826 MF741828 MF741829 MF741831 MF741833 MF741834 MF741835 MF741836 MG011343 MG011352 MG011353 MG011355 MF679172 MG366880 MG912608 MH306207 MH371127 MH432120 MG757593 MG757594 MG757595 MG757596 MG757598 MG757599 MG757600 MG757601 MG757602 MG757603 MH029552 MK129253 MH978888 MK462248 MH259486 MK858156 MK858158 MK858163 AHI48572.1 AHI48662.1 AHY22525.1 AHY22535.1 AID50418.1 AID55092.1 AJD81451.1 AKA63509.1 AKJ80137.2 AKK52582.1 AKK52592.1 AKK52602.1 AKK52612.1 AKL80593.1 AKL80604.1 AKL80615.1 AKN11071.1 AKQ21055.1 AKQ21064.1 AKQ21073.1 ALA49440.1 ALA49825.1 ALA49847.1 ALA49858.1 ALA49869.1 ALA49880.1 ALA49891.1 ALA49913.1 ALA49924.1 ALA49935.1 ALA49979.1 ALA49990.1 ALA50001.1 ALA50012.1 ALA50023.1 ALA50056.1 ALJ54444.1 ALJ54445.1 ALJ54447.1 ALJ54449.1 ALJ54454.1 ALJ54457.1 ALJ54459.1 ALJ54460.1 ALJ54462.1 ALJ54464.1 ALJ54469.1 ALJ54470.1 ALJ54473.1 ALJ54476.1 ALJ54482.1 ALJ54485.1 ALJ54487.1 ALJ54489.1 ALJ54492.1 ALJ54494.1 ALJ54498.1 ALJ54499.1 ALJ54503.1 ALJ54504.1 ALJ54505.1 ALJ54506.1 ALJ54507.1 ALJ54509.1 ALJ54510.1 ALJ54511.1 ALJ54513.1 ALJ54514.1 ALJ54515.1 ALJ54519.1 ALK80242.1 ALT66802.1 ALT66813.1 ALT66824.1 ALW82636.1 ALW82647.1 ALW82658.1 ALW82669.1 ALW82680.1 ALW82702.1 ALW82709.1 ALW82720.1 ALW82731.1 AMQ49015.1 AMQ49026.1 AMQ49037.1 AMQ49048.1 AMQ49059.1 AMQ49070.1 ANF29162.1 ANI69911.1 AQZ41311.1 AQZ41332.1 AQZ41343.1 AQZ41354.1 AQZ41365.1 AQZ41376.1 ASU89921.1 ASU89933.1 ASU90021.1 ASU90032.1 ASU90054.1 ASU90065.1 ASU90109.1 ASU90120.1 ASU90131.1 ASU90153.1 ASU90208.1 ASU90263.1 ASU90285.1 ASU90295.1 ASU90351.1 ASU90461.1 ASU90472.1 ASU90483.1 ASU90494.1 ASU90560.1 ASU90582.1 ASU90615.1 ASU90648.1 ASU90659.1 ASU90670.1 ASU90692.1 ASU90703.1 ASU90714.1 ASU90736.1 ASU90758.1 ASU90835.1 ASU90868.1 ASU90890.1 ASU90912.1 ASU90923.1 ASU90934.1 ASU90945.1 ASU90956.1 ASU90988.1 ASU90999.1 ASU91021.1 ASU91032.1 ASU91054.1 ASU91065.1 ASU91098.1 ASY99778.1 ASY99789.1 ASY99800.1 ASY99820.1 ASY99831.1 ASY99853.1 ASY99873.1 ASY99884.1 ASY99895.1 ASY99906.1 ATG84712.1 ATG84811.1 ATG84822.1 ATG84844.1 ATW75478.1 ATY74381.1 AVK87461.1 AWM99582.1 AXG21643.1 AXG21665.1 AXN73393.1 AXN73404.1 AXN73415.1 AXN73426.1 AXN73448.1 AXN73459.1 AXN73470.1 AXN73481.1 AXN73492.1 AXN73503.1 AYO86699.1 AZK15900.1 QBF44115.1 QBF80510.1 QCI31480.1 QCQ28828.1 QCQ28830.1 QCQ28835.1 MK462253 MK462254 MK462255 QBF80565.1 QBF80576.1 QBF80587.1 MF598600 MF598678 ASU89988.1 ASU90846.1 MF598632 ASU90340.1 MF598679 ASU90857.1 KT806054 ALW82742.1 MF598623 ASU90241.1 KT805994 ALJ54474.1 MF598651 ASU90549.1 KX154693 ANF29261.1 MN403102 QEU56412.1 KT868872 KT868876 ALK80261.1 ALK80301.1
Proteomes
UP000007451
UP000148908
UP000164987
UP000143769
UP000100668
UP000124656
+ More
UP000007449 UP000107989 UP000163538 UP000006574 UP000123539 UP000110642 UP000104899 UP000145271 UP000167218 UP000138672 UP000162302 UP000149108 UP000146591 UP000098431 UP000104003 UP000112676 UP000126955 UP000128936 UP000136645 UP000137287 UP000146604 UP000151733 UP000159793 UP000148810 UP000113143 UP000152659 UP000140430 UP000154518 UP000134703 UP000157238 UP000119765 UP000120438 UP000135014 UP000148657 UP000159930 UP000117213 UP000118772 UP000104220 UP000112550 UP000112279 UP000095864 UP000096875 UP000102966 UP000104540 UP000109666 UP000112022 UP000115322 UP000118563 UP000119223 UP000121965 UP000122049 UP000125198 UP000128184 UP000128186 UP000128651 UP000142239 UP000144056 UP000144472 UP000147538 UP000151795 UP000153269 UP000157492 UP000158935 UP000159306 UP000161565 UP000163035 UP000163895 UP000170921 UP000171516 UP000171892 UP000174054 UP000174558 UP000180347 UP000180348 UP000171389
UP000007449 UP000107989 UP000163538 UP000006574 UP000123539 UP000110642 UP000104899 UP000145271 UP000167218 UP000138672 UP000162302 UP000149108 UP000146591 UP000098431 UP000104003 UP000112676 UP000126955 UP000128936 UP000136645 UP000137287 UP000146604 UP000151733 UP000159793 UP000148810 UP000113143 UP000152659 UP000140430 UP000154518 UP000134703 UP000157238 UP000119765 UP000120438 UP000135014 UP000148657 UP000159930 UP000117213 UP000118772 UP000104220 UP000112550 UP000112279 UP000095864 UP000096875 UP000102966 UP000104540 UP000109666 UP000112022 UP000115322 UP000118563 UP000119223 UP000121965 UP000122049 UP000125198 UP000128184 UP000128186 UP000128651 UP000142239 UP000144056 UP000144472 UP000147538 UP000151795 UP000153269 UP000157492 UP000158935 UP000159306 UP000161565 UP000163035 UP000163895 UP000170921 UP000171516 UP000171892 UP000174054 UP000174558 UP000180347 UP000180348 UP000171389
Interpro
SUPFAM
SSF143587
SSF143587
Gene 3D
ProteinModelPortal
A3EXD0
S4WZQ4
S4X422
S4WWR8
S4WZR3
S4WWS3
+ More
A0A0U1WHM0 S4WZR0 S4X278 S4X284 S4WWR5 A3EXD9 A3EXE8 A3EXF7 S4WYF2 S4WYE5 S4X419 S4WYE8 S4X425 S4X276 Q0Q4F7 A0A2I6PIW5 A0A2I6PIX8 A0A023Y9K3 A0A2I4R8V9 A0A2I4R888 A0A2R4KP93 A0A2R4KP86 A0A5H2WTJ3 A0A1Z1W230 A0A1Z1W244 S4WYD2 Q0Q4F2 A0A1Z1W227 S4WWQ3 S4WZQ1 A0A166ZLN4 S4WYD6 A3EXA3 A3EX94 A3EXC1 A3EXB2 A0A0U1WJZ6 A0A140AZ05 A0A0A0Q4U2 A0A0P0HTL4 A0A291I6G6 A0A0U2J5T6 A0A0U2INY2 A0A126G7Z3 A0A0U2MGJ8 A0A0U2MS80 A0A2H1YPP7 A0A0P0GPV1 A0A140AQ46 A0A0U2NTX5 A0A0U4AGH8 A0A0P0HJE7 A0A2D0YM48 A0A2D0YGK8 A0A0P0HBE6 A0A0U2P9C9 A0A0A0Q7F8 A0A0U2NE92 A0A0A0Q7G4 A0A2D0YIQ0 A0A1D6Y6X4 A0A0U2P188 A0A0U2J5U2 A0A2D0YCT4 T2B9T0 A0A0U2GMX5 T2B999 A0A0U2GLS2 W5ZZF5 A0A0U2MS90 A0A0U2NXW5 A0A3T0I8D8 A0A0G3U597 A0A2D0YCE0 R9UCW7 A0A1C8EAZ2 A0A0U2GPS7 A0A0U2P195 A0A291I6B7 A0A3S7R2I2 K0BRG7 A0A384VXU1 W6A028 A0A411PBL8 A0A2D0YNJ0 A0A2D0YPZ8 A0A2D0YAP4 A0A0U3SQE0 A0A2D0Y380 A0A0U2NE84 A0A2D0YDA3 A0A1W5LGU2 A0A5J6ED89 A0A140AZ15
A0A0U1WHM0 S4WZR0 S4X278 S4X284 S4WWR5 A3EXD9 A3EXE8 A3EXF7 S4WYF2 S4WYE5 S4X419 S4WYE8 S4X425 S4X276 Q0Q4F7 A0A2I6PIW5 A0A2I6PIX8 A0A023Y9K3 A0A2I4R8V9 A0A2I4R888 A0A2R4KP93 A0A2R4KP86 A0A5H2WTJ3 A0A1Z1W230 A0A1Z1W244 S4WYD2 Q0Q4F2 A0A1Z1W227 S4WWQ3 S4WZQ1 A0A166ZLN4 S4WYD6 A3EXA3 A3EX94 A3EXC1 A3EXB2 A0A0U1WJZ6 A0A140AZ05 A0A0A0Q4U2 A0A0P0HTL4 A0A291I6G6 A0A0U2J5T6 A0A0U2INY2 A0A126G7Z3 A0A0U2MGJ8 A0A0U2MS80 A0A2H1YPP7 A0A0P0GPV1 A0A140AQ46 A0A0U2NTX5 A0A0U4AGH8 A0A0P0HJE7 A0A2D0YM48 A0A2D0YGK8 A0A0P0HBE6 A0A0U2P9C9 A0A0A0Q7F8 A0A0U2NE92 A0A0A0Q7G4 A0A2D0YIQ0 A0A1D6Y6X4 A0A0U2P188 A0A0U2J5U2 A0A2D0YCT4 T2B9T0 A0A0U2GMX5 T2B999 A0A0U2GLS2 W5ZZF5 A0A0U2MS90 A0A0U2NXW5 A0A3T0I8D8 A0A0G3U597 A0A2D0YCE0 R9UCW7 A0A1C8EAZ2 A0A0U2GPS7 A0A0U2P195 A0A291I6B7 A0A3S7R2I2 K0BRG7 A0A384VXU1 W6A028 A0A411PBL8 A0A2D0YNJ0 A0A2D0YPZ8 A0A2D0YAP4 A0A0U3SQE0 A0A2D0Y380 A0A0U2NE84 A0A2D0YDA3 A0A1W5LGU2 A0A5J6ED89 A0A140AZ15
PDB
5X5F
E-value=0,
Score=4117
Ontologies
KEGG
GO
GO:0019031 C:viral envelope
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0016021 C:integral component of membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0020002 C:host cell plasma membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0061025 P:membrane fusion
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0016021 C:integral component of membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0020002 C:host cell plasma membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0061025 P:membrane fusion
Subcellular Location
From MSLVP
Host nucleus > host nucleolus. Host cytoplasm. Secreted.
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
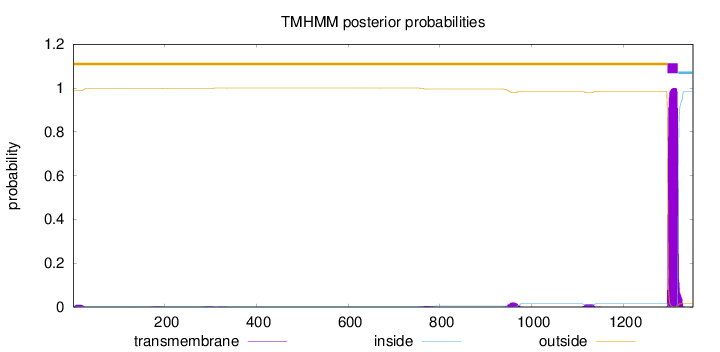
Topology
Length:
1352
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.39129
Exp number, first 60 AAs:
0.18076
Total prob of N-in:
0.01243
outside
1 - 1296
TMhelix
1297 - 1319
inside
1320 - 1352
Population Genetic Test Statistics
Pi
0.5279428
Theta
0.6215625
Tajima's D
-0.6558559
CLR
9.312744
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 |
Protein |
YP_001039962.1 |
|
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.