Strain
CHINA_BAT_2005_NC_009657
(Region: China; Strain: Scotophilus bat coronavirus 512, complete genome.; Date: 2005)
Gene
spike protein
Description
Annotated in NCBI,
spike protein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGAAATATACACTTTTATTTTGTGTAGTTTTTGCTACGGTGTCTTTTGGCTTTGCCGATAATGAGCGCTGTAACAAAACTGTCAACCTTACTAGGTTATTTTCTAAGTTTGATATACAACCACCATCTCAAGTTGTTTTAGCTGGTCTTTTACCAAATCAGACAGCACAATGGAAGTGCACCACAGAAACTAACAAAAGAGATGAGGGTGTTGGTGTTAAGGGTGTCTTTCTTAGCTATGTGAGTTCTGGGCGTGGGTTTACGATAGGGGTTTCTCAATATAACTTTGACCCTAGTACTTACCAGCTTTACTTGCACAGAGATACAAATGGCAACTCTAATGCTTTTGCGTATTTGCGCATATGCAAGTGGCCCAGCAAGAAATGGTTACAATCTACATCCAACATGGATACCAGTGGTAGGTTTTGTTTGGTCAACAAGAAAATACCAGCTGCGTTTACTGATCATGCCAATATGGTTGTCGGTATAACATGGGACCAAGATCGGGTCACTTTTTACACTGACAAAGTCTACCATTTTTATGTCCCAAACAACCGCTGGTCACGTGTGGTGTCGTGGTGTAGTGCTGCGGATAGTTGTGCTATGCAATATATTAACAGTACTATCTATTACAATCTTAATGTAACAACACCAGGGCCTGGTGGTATCACTTATTCTGTTTGTACAAAACATTGTACTGGTTTGGCAGACAATGTTTTTTCCACAGATCAGGGTGGCCACATACCACCTATTTTTCCGTACAACAATTGGTTCTTGTTGACTAACACATCCACGTTGGTACAGGGTGTTACTCGTGTGTTTCAACCATTTCTTGTCAATTGCTTGGTTGCATTGCCTAAGTTGCAAGGTCTTACTACCACTCTGTCTTTTGACTCACCACTTAATGTGCCTGGGTTTTCCTGTAACGGCGCCAATGGTTCTAGCTCAGCGGAAGCCTTTCGTTTTAACGTCAATGATACTAAGTTGTTTGTTGGTGCTGGCGCTGTTACATTGAACACCGTCGATGGTGTTAATGTTTCTATTGTGTGCTCCAATAATGCAACACAGCCCACTAGGTCAAACAACTTGCAGGAAGACCTGCCTTACTATTGCTTCACTAACACTAGTAGCGGCACTAATCACACTGTTAAGTTTCTTTCAGTTTTCCCGCCAATCATTCGTGAGTTTGTGATCACCAAATATGGCAATGTCTATGTTAATGGCTATATCTATTTGAGAACTAGACCATTGACAGCCGTGCACTTGAACGCATCCTCTCATTCGCAGGACGTAGCAGGGTTTTGGACTATTGCCGCCACAAACTTCACGGATGTGCTTGTTGAGGTGAACAACACAGGCATTCAGAGGTTGTTGTATTGTGACACGCCTGAAAACAGTGTCAAATGTTCACAACTCTCTTTTGAACTGGAGGACGGGTTTTATTCCATGACTGCAGATAATGTTTATGCAGTAACTAAGCCCCACACGTTTGTGACTTTGCCCACGTTTAATGACCATGGGTTCGTTAATGTTACTGTGGGTGGTAACTTTGACAGTTCATACCCACCAAAGTTCACTGCTAATGGCACCTTAGTTAATAACGGCACTGTGGTGTGTGTCACTTCTAATCAGTTCACCCTTAGACACGACTTTATGGTAGGTTATTCTGCTGATATGCGTAAGGGTATATTTGAGTACTCTAGTACATGCCCTTTCAATAGAGAAACTATCAATAACTACCTTACGTTTGGTCGTATTTGTTTCTCTACTTCACCGGCGGACGGTGCTTGCGAATTGAAGTACTATGTTTGGAACACCATTGGAGCCGTTTCACACCTGGCTGGCACCTTGTATGTTCAACATACAAAGGGTGACATAATAACTGGTACACCCAAACCATTGCAGGGTTTGAATGACATTTCTGAATTGCACCTAGACACGTGCACCACTTACACCATTTATGGTTTTAGGGGTGACGGTGTTATTAGGTTGACCAATCAAACTTTCTTGTCAGGTGTCTACTACACTTCAGAGAGTGGTCAGTTATTAGCTTTTAAGAATGTCACTACAGGGCAGATTTATTCTGTTACACCCTGCCAACTGGTTCAGCAGGTTGCTTTTGTTGAGGATAGGATTGTTGGCGTCATTAGTAGTGCTAATAATACTGGGTTCTTTAATTCCACAAGAACATTTCCAGGCTTCTATTATCACTCTAATGACACCACCAATTGCACCTCACCAAGACTTGTTTACTCTAATATAGGTGTTTGTACTAGTGGTGCCATAGGTTTGCTGTCTCCTAAAGCTGCACAACCTCAGGTTCAACCCATGTTCCAGGGTAATATTAGTATCCCTACTAATTTTACTATGAGTGTGCGCACTGAGTATATACAGTTGTTTAACAAACCCGTTTCTGTAGACTGCGCAATGTATGTCTGCAATGGTAATGACCGTTGTAAGCAATTGTTGTCTCAGTACACTTCAGCATGCAAGAACATAGAATCTGCGCTGCAGCTCAGCGCAAGGTTGGAATCAATGGAGGTTAACTCTATGTTGACAGTTTCAGATGAGGCACTTAAGCTTGCCACTATAAGCCAATTTCCTGGTGGTGGTTATAATTTTACCAATATTCTTCCAGCAAATCCTGGTGCTAGGTCAGTTATTGAAGACATTTTGTTCGATAAAGTTGTCACTAGTGGTTTGGGCACAGTTGATGAAGATTATAAACGCTGCAGTAATGGACTGTCTATTGCAGATTTAGCTTGTGCGCAGCACTATAACGGCATTATGGTGTTGCCGGGTGTTGCGGACTGGGAAAAGGTCCATATGTACTCGGCTTCACTTGTCGGTGGTATGACCTTAGGTGGTATCACTTCTGCTGCGGCTTTGCCTTTCTCATATGCAGTGCAGGCAAGACTTAATTATGTTGCACTACAGACCGACGTGCTGCAACGTAATCAACAAATGCTAGCCAATTCCTTTAATAGTGCTATTAGTAACATCACATTAGCTTTTGAGAGTGTCAATAACGCTATCTATCAAACTTCTGCTGGTTTGAATACGGTAGCAGAGGCACTTTCAAAAGTACAGGATGTTGTGAATGGTCAAGGAAATGCACTCAGTCAACTAACAGTCCAATTGCAGAATAATTTTCAAGCTATTTCCAATTCTATTGGTGACATTTATAGTAGGTTAGATCAGATAACTGCTGATGCGCAAGTTGACAGACTTATCACAGGTCGGCTTGCAGCTCTTAATGCCTTTGTTGCACAGTCACTTACCAAGTATGCAGAAGTGCAAGCTAGTAGGACATTGGCCAAGCAAAAGGTTAACGAGTGTGTTAAGTCACAGTCCCCCAGATACGGTTTCTGTGGTGATGAAGGGGAACATATTTTCTCACTCACCCAAGCTGCTCCACAGGGTCTGATGTTCCTACACACCGTTTTAGTACCTAATGGTTTTATTAACGTTACAGCAGTTACAGGTTTATGTGTTGATGAGACCATAGCTATGACATTACGTCAGAGTGGATTTGTCTTGTTTGTGCAAAATGGTAATTATCTCGTGTCACCGAGGAAAATGTTTGAACCTCGGAGACCTGAAGTTGCTGATTTTGTGCAAGTAAAAACATGCACGATTAGTTATGTTAACATCACCAATAACCAGTTGCCTGACATTATTCCAGATTATGTAGACGTTAATAAGACTATAGATGAGATTTTGGCCAACCTACCTAATAATACTGTGCCTGATTTGCCACTTGATGTCTTTAATCAAACATTTCTTAATCTCACTGGTGAGATTGCAGACCTTGAAGCGCGATCTGAATCCCTTAAAAACACATCAGAAGAACTTAGACAGTTGATCCAAAATATTAACAACACACTTGTAGACCTTCAGTGGCTTAATAGGGTTGAGACCTTTATTAAGTGGCCGTGGTACGTGTGGTTGGCTATTGTTATAGCTCTTATTTTGGTTGTTTCACTGCTTGTGTTCTGCTGTATATCTACAGGTTGTTGCGGTTGTTGCGGTTGTTGTGGTTCTTGTTTCTCAGGTTGTTGTCGTGGAACTAAACTTCAACATTACGAACCAATAGAAAAGGTTCATGTGCAATAA
Protein
MKYTLLFCVVFATVSFGFADNERCNKTVNLTRLFSKFDIQPPSQVVLAGLLPNQTAQWKCTTETNKRDEGVGVKGVFLSYVSSGRGFTIGVSQYNFDPSTYQLYLHRDTNGNSNAFAYLRICKWPSKKWLQSTSNMDTSGRFCLVNKKIPAAFTDHANMVVGITWDQDRVTFYTDKVYHFYVPNNRWSRVVSWCSAADSCAMQYINSTIYYNLNVTTPGPGGITYSVCTKHCTGLADNVFSTDQGGHIPPIFPYNNWFLLTNTSTLVQGVTRVFQPFLVNCLVALPKLQGLTTTLSFDSPLNVPGFSCNGANGSSSAEAFRFNVNDTKLFVGAGAVTLNTVDGVNVSIVCSNNATQPTRSNNLQEDLPYYCFTNTSSGTNHTVKFLSVFPPIIREFVITKYGNVYVNGYIYLRTRPLTAVHLNASSHSQDVAGFWTIAATNFTDVLVEVNNTGIQRLLYCDTPENSVKCSQLSFELEDGFYSMTADNVYAVTKPHTFVTLPTFNDHGFVNVTVGGNFDSSYPPKFTANGTLVNNGTVVCVTSNQFTLRHDFMVGYSADMRKGIFEYSSTCPFNRETINNYLTFGRICFSTSPADGACELKYYVWNTIGAVSHLAGTLYVQHTKGDIITGTPKPLQGLNDISELHLDTCTTYTIYGFRGDGVIRLTNQTFLSGVYYTSESGQLLAFKNVTTGQIYSVTPCQLVQQVAFVEDRIVGVISSANNTGFFNSTRTFPGFYYHSNDTTNCTSPRLVYSNIGVCTSGAIGLLSPKAAQPQVQPMFQGNISIPTNFTMSVRTEYIQLFNKPVSVDCAMYVCNGNDRCKQLLSQYTSACKNIESALQLSARLESMEVNSMLTVSDEALKLATISQFPGGGYNFTNILPANPGARSVIEDILFDKVVTSGLGTVDEDYKRCSNGLSIADLACAQHYNGIMVLPGVADWEKVHMYSASLVGGMTLGGITSAAALPFSYAVQARLNYVALQTDVLQRNQQMLANSFNSAISNITLAFESVNNAIYQTSAGLNTVAEALSKVQDVVNGQGNALSQLTVQLQNNFQAISNSIGDIYSRLDQITADAQVDRLITGRLAALNAFVAQSLTKYAEVQASRTLAKQKVNECVKSQSPRYGFCGDEGEHIFSLTQAAPQGLMFLHTVLVPNGFINVTAVTGLCVDETIAMTLRQSGFVLFVQNGNYLVSPRKMFEPRRPEVADFVQVKTCTISYVNITNNQLPDIIPDYVDVNKTIDEILANLPNNTVPDLPLDVFNQTFLNLTGEIADLEARSESLKNTSEELRQLIQNINNTLVDLQWLNRVETFIKWPWYVWLAIVIALILVVSLLVFCCISTGCCGCCGCCGSCFSGCCRGTKLQHYEPIEKVHVQ
Summary
Function
S1 region attaches the virion to the cell membrane by interacting with host ANPEP/aminopeptidase N, initiating the infection. Binding to the receptor probably induces conformational changes in the S glycoprotein unmasking the fusion peptide of S2 region and activating membranes fusion. S2 region belongs to the class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) regions assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer. During virus morphogenesis, found in a complex with M and HE proteins. Interacts with host ANPEP.
Miscellaneous
Bat coronavirus 512/2005 is highly similar to porcine epidemic diarrhea virus (PEDV).
Similarity
Belongs to the alphacoronaviruses spike protein family.
Keywords
Coiled coil
Glycoprotein
Host membrane
Host-virus interaction
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Virion
Virulence
Virus entry into host cell
3D-structure
Feature
chain Spike glycoprotein
Uniprot
Q0Q466
Q0Q4F6
A0A3G3NGJ7
A0A4Y6GL57
A0A4Y6GL18
A0A3G3NGM2
+ More
A0A3G3NHI3 Q0Q4F5 A0A3G3NGK2 A0A219TUS3 A0A3G3NHJ0 A0A3G3NGV3 A0A3G3NGN7 A0A3G3NH05 A0A4Y6GL52 A0A3G3NGW9 A0A3G3NHL7 A0A3G3NHH0 A0A3G3NHH9 A0A3G3NGX8 A0A3G3NGM9 A0A3G3NGQ4 A0A3G3NGG7 A0A3G3NGQ0 A0A3G3NGS7 A0A3G3NGT9 I1TLC3 A0A0U3HSZ7 Q0GJJ0 A0A0M4I404 G5D078 A0A2N8Z1W1 A0A0K0WTL0 Q91AV1 A0A239QPZ3 A0A238TMM2 A0A0U1VEL1 A0A1B3Q5X6 A0A1P8A7E4 A0A238TN08 A0A0H4AC20 A0A3G2C516 A0A0M5JV19 J7LKT6 A0A5A4M1V6 A0A075ECF0 A0A109QKG8 A0A239QNQ6 A0A0F6TKU6 A0A238TQ27 A0A5A4M032 A0A0H4A7A7 A0A2N8Z212 K4G1P9 A0A238TMM6 A0A2N8Z1U7 I2EBM8 A0A5A4LZX3 A0A0M4HL41 A0A075EC92 A0A0K1LUK6 A0A2R4H2S5 A0A5A4M1T6 A0A0K1LUM5 A0A076MKN4 A0A221HZH0 S4VA59 A0A089WZ38 A0A238TM74 A0A2U8QMX8 A0A219PSP4 A0A075EAW2 A0A5A4LZS9 A0A239QPT4 A0A238TMH6 A0A2U8QMX5 A0A0M4HL64 A0A125R587 A0A0H4AE20 I3WD89 A0A5A4LZS2 A0A0K2D763 W8Q548 A0A2N8Z1T8 A0A0K2D774 A0A2R4FC33 A0A0S3MR49 A0A2H5AC28 A0A238TPF5 A0A0A8LEG6 A0A1L3KJ30 A0A1L3GVA2 A0A0A8LF79 A0A5A4M655 A0A1C7A0X8 A0A5J6KLN8 A0A1L3GVC2 A0A5A4M3B5 A0A238TMZ1
A0A3G3NHI3 Q0Q4F5 A0A3G3NGK2 A0A219TUS3 A0A3G3NHJ0 A0A3G3NGV3 A0A3G3NGN7 A0A3G3NH05 A0A4Y6GL52 A0A3G3NGW9 A0A3G3NHL7 A0A3G3NHH0 A0A3G3NHH9 A0A3G3NGX8 A0A3G3NGM9 A0A3G3NGQ4 A0A3G3NGG7 A0A3G3NGQ0 A0A3G3NGS7 A0A3G3NGT9 I1TLC3 A0A0U3HSZ7 Q0GJJ0 A0A0M4I404 G5D078 A0A2N8Z1W1 A0A0K0WTL0 Q91AV1 A0A239QPZ3 A0A238TMM2 A0A0U1VEL1 A0A1B3Q5X6 A0A1P8A7E4 A0A238TN08 A0A0H4AC20 A0A3G2C516 A0A0M5JV19 J7LKT6 A0A5A4M1V6 A0A075ECF0 A0A109QKG8 A0A239QNQ6 A0A0F6TKU6 A0A238TQ27 A0A5A4M032 A0A0H4A7A7 A0A2N8Z212 K4G1P9 A0A238TMM6 A0A2N8Z1U7 I2EBM8 A0A5A4LZX3 A0A0M4HL41 A0A075EC92 A0A0K1LUK6 A0A2R4H2S5 A0A5A4M1T6 A0A0K1LUM5 A0A076MKN4 A0A221HZH0 S4VA59 A0A089WZ38 A0A238TM74 A0A2U8QMX8 A0A219PSP4 A0A075EAW2 A0A5A4LZS9 A0A239QPT4 A0A238TMH6 A0A2U8QMX5 A0A0M4HL64 A0A125R587 A0A0H4AE20 I3WD89 A0A5A4LZS2 A0A0K2D763 W8Q548 A0A2N8Z1T8 A0A0K2D774 A0A2R4FC33 A0A0S3MR49 A0A2H5AC28 A0A238TPF5 A0A0A8LEG6 A0A1L3KJ30 A0A1L3GVA2 A0A0A8LF79 A0A5A4M655 A0A1C7A0X8 A0A5J6KLN8 A0A1L3GVC2 A0A5A4M3B5 A0A238TMZ1
Pubmed
EMBL
DQ648858
DQ648791
ABG11963.1
MH687941
AYR18442.1
MK211371
+ More
QDF43800.1 MK211370 QDF43795.1 MH687934 AYR18400.1 MH687938 AYR18424.1 DQ648792 ABG11964.1 MH687948 AYR18481.1 KT346372 ANV78630.1 MH687954 AYR18513.1 MH687940 AYR18436.1 MH687951 AYR18499.1 MH687952 AYR18505.1 MK211369 QDF43790.1 MH687967 AYR18591.1 MH687964 AYR18573.1 MH687946 AYR18473.1 MH687950 AYR18493.1 MH687963 AYR18567.1 MH687945 AYR18467.1 MH687939 AYR18430.1 MH687936 MH687959 AYR18412.1 AYR18543.1 MH687949 AYR18487.1 MH687935 AYR18406.1 MH687962 AYR18561.1 JQ023161 AFE85962.1 KU297956 ALU34112.1 DQ862099 ABI30278.1 KT206205 ALD19741.1 JN547228 AEQ55004.1 LT906620 SNX30664.1 KP768390 AKS26489.1 AF353511 LT898439 LT900499 SNQ28038.1 SNT95659.1 LT898436 LT898435 SNQ28016.1 SNQ28045.1 KJ857477 AJD09600.1 KU985229 AOG30831.1 KU975401 AMR60838.1 LT898431 SNQ27978.1 KR296666 AKN45968.1 MG706122 AYM47397.1 KT206204 LT898418 LT898433 LT898441 ALD19740.1 SNQ27931.1 SNQ27997.1 SNQ28051.1 JX242462 AFR11480.1 MG198645 AXS76469.1 KJ645655 AID56787.1 KT428878 AMB20700.1 LT898413 LT900502 SNQ27899.1 SNT95671.1 KR011756 LT898416 LT898409 LT898410 LT898430 LT898446 LT900498 AKE47378.1 SNQ27923.1 SNQ27941.1 SNQ27961.1 SNQ27991.1 SNQ28064.1 SNT95653.1 LT898432 LT898443 SNQ27967.1 SNQ28057.1 MG198634 AXS76458.1 KR296682 AKN45984.1 LT906581 SNW64190.1 JQ979287 AFM55051.1 LT898423 SNQ27930.1 LT906582 SNW64184.1 JQ638923 AFJ97038.1 MG198640 AXS76464.1 KP399614 ALD19703.1 KJ645635 KJ645702 KJ645704 LC063846 AID56667.1 AID57069.1 AID57081.1 BAT33329.1 KR011121 AKU46226.1 MH006958 AVU05417.1 MG198635 AXS76459.1 KR011122 LT898415 LT898414 LT898411 LT898412 LT898425 LT898427 LT898426 LT898438 LT898444 LT898417 AKU46227.1 SNQ27893.1 SNQ27911.1 SNQ27917.1 SNQ27955.1 SNQ27973.1 SNQ27985.1 SNQ28010.1 SNQ28022.1 SNQ28076.1 SNQ28082.1 KJ645695 KJ645696 KM196109 KM392232 KR265759 KR265760 KM975738 KM975739 KM975740 KY619745 AID57027.1 AID57033.1 AIJ19447.1 AIN52136.1 AKJ21701.1 AKJ21707.1 AKP80610.1 AKP80617.1 AKP80624.1 ATJ03600.1 KY381581 LT898447 ASM46788.1 SNQ28088.1 KC787539 KP870120 AGO59789.1 ALA15713.1 KM403155 KM403156 KM403157 AIR95864.1 AIR95870.1 AIR95871.1 LT898421 SNQ27905.1 MH052686 AWM11504.1 KT591944 AMD11118.1 KJ645649 AID56751.1 MG198651 MG198664 AXS76475.1 AXS76488.1 LT900500 SNT95665.1 LT898440 SNQ28032.1 MH052680 AWM11503.1 KT206206 ALD19742.1 KT860508 AMD11124.1 KR296678 AKN45980.1 JQ239434 AFL02629.1 MG198641 AXS76465.1 KP870118 ALA15711.1 KJ399978 AHL38184.1 LT905450 SNU46336.1 KP870128 ALA15721.1 MF038017 AVT28076.1 LC063847 BAT33335.1 KY828991 AUG68851.1 LT898408 SNQ27887.1 LM645057 CDW77205.1 KX883635 APG77336.1 KY019624 APG31005.1 LM645058 CDW77213.1 MG198633 AXS76457.1 KM975741 AKP80631.1 MK648472 QEV88420.1 KY019623 APG30998.1 MG198659 AXS76483.1 LT898420 SNQ27949.1
QDF43800.1 MK211370 QDF43795.1 MH687934 AYR18400.1 MH687938 AYR18424.1 DQ648792 ABG11964.1 MH687948 AYR18481.1 KT346372 ANV78630.1 MH687954 AYR18513.1 MH687940 AYR18436.1 MH687951 AYR18499.1 MH687952 AYR18505.1 MK211369 QDF43790.1 MH687967 AYR18591.1 MH687964 AYR18573.1 MH687946 AYR18473.1 MH687950 AYR18493.1 MH687963 AYR18567.1 MH687945 AYR18467.1 MH687939 AYR18430.1 MH687936 MH687959 AYR18412.1 AYR18543.1 MH687949 AYR18487.1 MH687935 AYR18406.1 MH687962 AYR18561.1 JQ023161 AFE85962.1 KU297956 ALU34112.1 DQ862099 ABI30278.1 KT206205 ALD19741.1 JN547228 AEQ55004.1 LT906620 SNX30664.1 KP768390 AKS26489.1 AF353511 LT898439 LT900499 SNQ28038.1 SNT95659.1 LT898436 LT898435 SNQ28016.1 SNQ28045.1 KJ857477 AJD09600.1 KU985229 AOG30831.1 KU975401 AMR60838.1 LT898431 SNQ27978.1 KR296666 AKN45968.1 MG706122 AYM47397.1 KT206204 LT898418 LT898433 LT898441 ALD19740.1 SNQ27931.1 SNQ27997.1 SNQ28051.1 JX242462 AFR11480.1 MG198645 AXS76469.1 KJ645655 AID56787.1 KT428878 AMB20700.1 LT898413 LT900502 SNQ27899.1 SNT95671.1 KR011756 LT898416 LT898409 LT898410 LT898430 LT898446 LT900498 AKE47378.1 SNQ27923.1 SNQ27941.1 SNQ27961.1 SNQ27991.1 SNQ28064.1 SNT95653.1 LT898432 LT898443 SNQ27967.1 SNQ28057.1 MG198634 AXS76458.1 KR296682 AKN45984.1 LT906581 SNW64190.1 JQ979287 AFM55051.1 LT898423 SNQ27930.1 LT906582 SNW64184.1 JQ638923 AFJ97038.1 MG198640 AXS76464.1 KP399614 ALD19703.1 KJ645635 KJ645702 KJ645704 LC063846 AID56667.1 AID57069.1 AID57081.1 BAT33329.1 KR011121 AKU46226.1 MH006958 AVU05417.1 MG198635 AXS76459.1 KR011122 LT898415 LT898414 LT898411 LT898412 LT898425 LT898427 LT898426 LT898438 LT898444 LT898417 AKU46227.1 SNQ27893.1 SNQ27911.1 SNQ27917.1 SNQ27955.1 SNQ27973.1 SNQ27985.1 SNQ28010.1 SNQ28022.1 SNQ28076.1 SNQ28082.1 KJ645695 KJ645696 KM196109 KM392232 KR265759 KR265760 KM975738 KM975739 KM975740 KY619745 AID57027.1 AID57033.1 AIJ19447.1 AIN52136.1 AKJ21701.1 AKJ21707.1 AKP80610.1 AKP80617.1 AKP80624.1 ATJ03600.1 KY381581 LT898447 ASM46788.1 SNQ28088.1 KC787539 KP870120 AGO59789.1 ALA15713.1 KM403155 KM403156 KM403157 AIR95864.1 AIR95870.1 AIR95871.1 LT898421 SNQ27905.1 MH052686 AWM11504.1 KT591944 AMD11118.1 KJ645649 AID56751.1 MG198651 MG198664 AXS76475.1 AXS76488.1 LT900500 SNT95665.1 LT898440 SNQ28032.1 MH052680 AWM11503.1 KT206206 ALD19742.1 KT860508 AMD11124.1 KR296678 AKN45980.1 JQ239434 AFL02629.1 MG198641 AXS76465.1 KP870118 ALA15711.1 KJ399978 AHL38184.1 LT905450 SNU46336.1 KP870128 ALA15721.1 MF038017 AVT28076.1 LC063847 BAT33335.1 KY828991 AUG68851.1 LT898408 SNQ27887.1 LM645057 CDW77205.1 KX883635 APG77336.1 KY019624 APG31005.1 LM645058 CDW77213.1 MG198633 AXS76457.1 KM975741 AKP80631.1 MK648472 QEV88420.1 KY019623 APG30998.1 MG198659 AXS76483.1 LT898420 SNQ27949.1
Proteomes
UP000113079
UP000122353
UP000132054
UP000095955
UP000276154
UP000008159
+ More
UP000269355 UP000274124 UP000278990 UP000280596 UP000267609 UP000271078 UP000274992 UP000278554 UP000151262 UP000270497 UP000274353 UP000100204 UP000267271 UP000276298 UP000277020 UP000279022 UP000280517 UP000281047 UP000272790 UP000276964 UP000269222 UP000277354 UP000274194 UP000122852 UP000136506 UP000147669 UP000153418 UP000270191 UP000270545 UP000273819 UP000274834 UP000275549 UP000275656 UP000275831 UP000276052 UP000278039 UP000278495 UP000096695 UP000120524 UP000123161 UP000154382 UP000154704 UP000271135 UP000109281 UP000268721 UP000161506 UP000271448 UP000267306 UP000173775 UP000107011 UP000278337 UP000107825 UP000277927 UP000138211 UP000155002 UP000269867
UP000269355 UP000274124 UP000278990 UP000280596 UP000267609 UP000271078 UP000274992 UP000278554 UP000151262 UP000270497 UP000274353 UP000100204 UP000267271 UP000276298 UP000277020 UP000279022 UP000280517 UP000281047 UP000272790 UP000276964 UP000269222 UP000277354 UP000274194 UP000122852 UP000136506 UP000147669 UP000153418 UP000270191 UP000270545 UP000273819 UP000274834 UP000275549 UP000275656 UP000275831 UP000276052 UP000278039 UP000278495 UP000096695 UP000120524 UP000123161 UP000154382 UP000154704 UP000271135 UP000109281 UP000268721 UP000161506 UP000271448 UP000267306 UP000173775 UP000107011 UP000278337 UP000107825 UP000277927 UP000138211 UP000155002 UP000269867
ProteinModelPortal
Q0Q466
Q0Q4F6
A0A3G3NGJ7
A0A4Y6GL57
A0A4Y6GL18
A0A3G3NGM2
+ More
A0A3G3NHI3 Q0Q4F5 A0A3G3NGK2 A0A219TUS3 A0A3G3NHJ0 A0A3G3NGV3 A0A3G3NGN7 A0A3G3NH05 A0A4Y6GL52 A0A3G3NGW9 A0A3G3NHL7 A0A3G3NHH0 A0A3G3NHH9 A0A3G3NGX8 A0A3G3NGM9 A0A3G3NGQ4 A0A3G3NGG7 A0A3G3NGQ0 A0A3G3NGS7 A0A3G3NGT9 I1TLC3 A0A0U3HSZ7 Q0GJJ0 A0A0M4I404 G5D078 A0A2N8Z1W1 A0A0K0WTL0 Q91AV1 A0A239QPZ3 A0A238TMM2 A0A0U1VEL1 A0A1B3Q5X6 A0A1P8A7E4 A0A238TN08 A0A0H4AC20 A0A3G2C516 A0A0M5JV19 J7LKT6 A0A5A4M1V6 A0A075ECF0 A0A109QKG8 A0A239QNQ6 A0A0F6TKU6 A0A238TQ27 A0A5A4M032 A0A0H4A7A7 A0A2N8Z212 K4G1P9 A0A238TMM6 A0A2N8Z1U7 I2EBM8 A0A5A4LZX3 A0A0M4HL41 A0A075EC92 A0A0K1LUK6 A0A2R4H2S5 A0A5A4M1T6 A0A0K1LUM5 A0A076MKN4 A0A221HZH0 S4VA59 A0A089WZ38 A0A238TM74 A0A2U8QMX8 A0A219PSP4 A0A075EAW2 A0A5A4LZS9 A0A239QPT4 A0A238TMH6 A0A2U8QMX5 A0A0M4HL64 A0A125R587 A0A0H4AE20 I3WD89 A0A5A4LZS2 A0A0K2D763 W8Q548 A0A2N8Z1T8 A0A0K2D774 A0A2R4FC33 A0A0S3MR49 A0A2H5AC28 A0A238TPF5 A0A0A8LEG6 A0A1L3KJ30 A0A1L3GVA2 A0A0A8LF79 A0A5A4M655 A0A1C7A0X8 A0A5J6KLN8 A0A1L3GVC2 A0A5A4M3B5 A0A238TMZ1
A0A3G3NHI3 Q0Q4F5 A0A3G3NGK2 A0A219TUS3 A0A3G3NHJ0 A0A3G3NGV3 A0A3G3NGN7 A0A3G3NH05 A0A4Y6GL52 A0A3G3NGW9 A0A3G3NHL7 A0A3G3NHH0 A0A3G3NHH9 A0A3G3NGX8 A0A3G3NGM9 A0A3G3NGQ4 A0A3G3NGG7 A0A3G3NGQ0 A0A3G3NGS7 A0A3G3NGT9 I1TLC3 A0A0U3HSZ7 Q0GJJ0 A0A0M4I404 G5D078 A0A2N8Z1W1 A0A0K0WTL0 Q91AV1 A0A239QPZ3 A0A238TMM2 A0A0U1VEL1 A0A1B3Q5X6 A0A1P8A7E4 A0A238TN08 A0A0H4AC20 A0A3G2C516 A0A0M5JV19 J7LKT6 A0A5A4M1V6 A0A075ECF0 A0A109QKG8 A0A239QNQ6 A0A0F6TKU6 A0A238TQ27 A0A5A4M032 A0A0H4A7A7 A0A2N8Z212 K4G1P9 A0A238TMM6 A0A2N8Z1U7 I2EBM8 A0A5A4LZX3 A0A0M4HL41 A0A075EC92 A0A0K1LUK6 A0A2R4H2S5 A0A5A4M1T6 A0A0K1LUM5 A0A076MKN4 A0A221HZH0 S4VA59 A0A089WZ38 A0A238TM74 A0A2U8QMX8 A0A219PSP4 A0A075EAW2 A0A5A4LZS9 A0A239QPT4 A0A238TMH6 A0A2U8QMX5 A0A0M4HL64 A0A125R587 A0A0H4AE20 I3WD89 A0A5A4LZS2 A0A0K2D763 W8Q548 A0A2N8Z1T8 A0A0K2D774 A0A2R4FC33 A0A0S3MR49 A0A2H5AC28 A0A238TPF5 A0A0A8LEG6 A0A1L3KJ30 A0A1L3GVA2 A0A0A8LF79 A0A5A4M655 A0A1C7A0X8 A0A5J6KLN8 A0A1L3GVC2 A0A5A4M3B5 A0A238TMZ1
PDB
6U7K
E-value=0,
Score=4042
Ontologies
KEGG
GO
GO:0016021 C:integral component of membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019031 C:viral envelope
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019031 C:viral envelope
GO:0019064 P:fusion of virus membrane with host plasma membrane
Subcellular Location
From MSLVP
Host nucleus. Host cytoplasm.
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
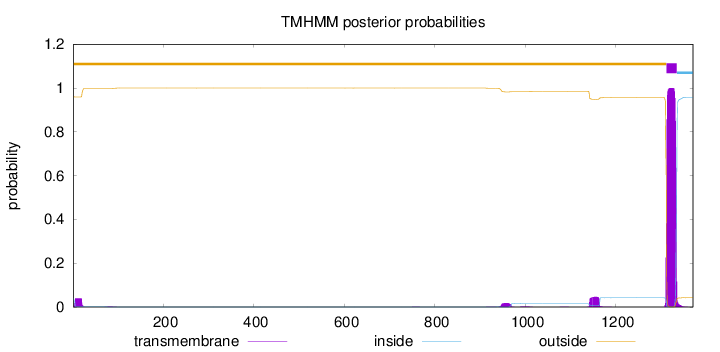
Topology
Length:
1371
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.85015
Exp number, first 60 AAs:
0.676359999999999
Total prob of N-in:
0.04045
outside
1 - 1312
TMhelix
1313 - 1335
inside
1336 - 1371
Population Genetic Test Statistics
Pi
0.3030282
Theta
0.5382972
Tajima's D
-1.677307
CLR
6.07
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 |
Protein |
YP_001351684.1 |
|
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.