Strain
CHINA_BAT_2005_NC_009657
(Region: China; Strain: Scotophilus bat coronavirus 512, complete genome.; Date: 2005)
Gene
putative ORF3
Description
Annotated in NCBI,
putative ORF3
Location
GenBank Accession
Full name
Non-structural protein 3
Alternative Name
Accessory protein 3a
Sequence
CDS
ATGTTTCTTGGTCTGTTCCAGTATACTATTGATACTGCAGTTGAGCACACTGTAGAACATGCTAACTTGTCCCAAGAAGAGGCTTTGATGTTGGAAGAAAACATCGTTCCTCTGAGACAAGCTACACATGTTACTGGATTTTTGCTCACCAGTGTTTTTGTTTACTTCTTTGCACTGTTTAAGGCTTCAAGCTACAAACGTAATTTGCTGCTATTTTTAGCACGTTTGTTAGCTTTATTAATTTATGCACCCATTTTAATATTTTGTGGTGCATACTTGGACGCTTTTATAGTAGTCGCAACATTGACTTCTCGTCTATTGTTTTTGACCTACTACTCATGGCGTTATAAAACTTATAAATTTCTTATTTACAACTCTTCCACACTTATGTTTTTACATGGTCATGCCAATTATTATAATGGCAGGCCCTATGTAATGCTTGAAGGTGGAAGCCATTACGTCACATTGGGTACTGATATAGTACCATTCGTCAGCCGAAGTAATCTCTATCTTGCCATTCGTGGTAGTGCTGAGTCAGATATCCAACTGTTGAGAACTGTCGAGTTGTTAGATGGTAATTACCTCTACATTTTCTCCAGTTGTCAAGTCGTTGGTGTTACTAATTCAGGTTTTGAGGAGATTCAACTAGACGAATATGCTACAATTAGTGAATGA
Protein
MFLGLFQYTIDTAVEHTVEHANLSQEEALMLEENIVPLRQATHVTGFLLTSVFVYFFALFKASSYKRNLLLFLARLLALLIYAPILIFCGAYLDAFIVVATLTSRLLFLTYYSWRYKTYKFLIYNSSTLMFLHGHANYYNGRPYVMLEGGSHYVTLGTDIVPFVSRSNLYLAIRGSAESDIQLLRTVELLDGNYLYIFSSCQVVGVTNSGFEEIQLDEYATISE
Summary
Miscellaneous
Bat coronavirus 512/2005 is highly similar to porcine epidemic diarrhea virus (PEDV).
Keywords
Host membrane
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Non-structural protein 3
Uniprot
A0A3G3NHP3
Q0Q465
A0A3G3NGB2
A0A3G3NHK5
A0A3G3NHE7
A0A3G3NGZ8
+ More
A0A219TU70 A0A3G3NGT8 A0A3G3NH43 A0A3G3NHL9 A0A3G3NGG5 A0A3G3NGW6 A0A3G3NHB8 A0A3G3NGZ1 A0A3G3NGE6 A0A3G3NHL5 A0A3G3NGF5 A0A3G3NGX3 A0A3G3NH25 A0A3G3NGJ8 A0A3G3NGI5 A0A5J6DDY7 A0A4Y6A773 D3XNW8 D3XNW4 A0A0U2UAH2 A0A0B5INN1 A0A411NPC9 A0A411NPH2 A0A4D6P035 A0A411NPJ9 A0A411NPE4 A0A1S6XZP4 A0A411NPH9 A0A411NPC3 A0A411NPC0 A0A411NPD4 A0A411NPH4 A0A411NPJ0 A0A411NPC1 A0A411NPK3 A0A411NPC8 A0A411NPJ1 A0A411NPK4 A0A411NPG1 A0A411NPI9 A0A411NPL8 A0A411NPB7 A0A411NPJ4 A0A411NPF5 A0A411NPH6 A0A411NPK5 A0A411NPM8 A0A411NPA8 A0A411NPE6 A0A411NPG9 A0A411NPH5 A0A411NPE5 A0A411NPD8 A0A411NPF3 A0A411NPK9 A0A411NPF2 A0A411NPF0 A0A411NPD9 A0A411NPJ5 A0A411NPI6 A0A0E3TJV2 A0A0U1VEK2 X5CC42 K4P5B0 L0CLX1 A0A2Z4N5X0 A0A190LBQ0 A0A0U1YH23 A0A167NFX0 A0A3G3C9B3 I3QJZ9 A0A160EEF0 L7Y082 S0API7 A0A411NPF9 A0A5J6VDZ1 A0A411NPF1 A0A097I514 A0A167NFF4 A0A097I537 A0A160EDB4 A0A238TPN3 A0A4D6FWV9 A0A0U2UEC2 A0A185QEM7 A0A0U2LMD3 A0A286R3Q8 A0A0B5J0Z8 A0A3S9VME3 A0A411NPD3 A0A160EDC3 U6A623 A0A190LBM1 A0A1Y0D9J8
A0A219TU70 A0A3G3NGT8 A0A3G3NH43 A0A3G3NHL9 A0A3G3NGG5 A0A3G3NGW6 A0A3G3NHB8 A0A3G3NGZ1 A0A3G3NGE6 A0A3G3NHL5 A0A3G3NGF5 A0A3G3NGX3 A0A3G3NH25 A0A3G3NGJ8 A0A3G3NGI5 A0A5J6DDY7 A0A4Y6A773 D3XNW8 D3XNW4 A0A0U2UAH2 A0A0B5INN1 A0A411NPC9 A0A411NPH2 A0A4D6P035 A0A411NPJ9 A0A411NPE4 A0A1S6XZP4 A0A411NPH9 A0A411NPC3 A0A411NPC0 A0A411NPD4 A0A411NPH4 A0A411NPJ0 A0A411NPC1 A0A411NPK3 A0A411NPC8 A0A411NPJ1 A0A411NPK4 A0A411NPG1 A0A411NPI9 A0A411NPL8 A0A411NPB7 A0A411NPJ4 A0A411NPF5 A0A411NPH6 A0A411NPK5 A0A411NPM8 A0A411NPA8 A0A411NPE6 A0A411NPG9 A0A411NPH5 A0A411NPE5 A0A411NPD8 A0A411NPF3 A0A411NPK9 A0A411NPF2 A0A411NPF0 A0A411NPD9 A0A411NPJ5 A0A411NPI6 A0A0E3TJV2 A0A0U1VEK2 X5CC42 K4P5B0 L0CLX1 A0A2Z4N5X0 A0A190LBQ0 A0A0U1YH23 A0A167NFX0 A0A3G3C9B3 I3QJZ9 A0A160EEF0 L7Y082 S0API7 A0A411NPF9 A0A5J6VDZ1 A0A411NPF1 A0A097I514 A0A167NFF4 A0A097I537 A0A160EDB4 A0A238TPN3 A0A4D6FWV9 A0A0U2UEC2 A0A185QEM7 A0A0U2LMD3 A0A286R3Q8 A0A0B5J0Z8 A0A3S9VME3 A0A411NPD3 A0A160EDC3 U6A623 A0A190LBM1 A0A1Y0D9J8
EMBL
MH687945
MH687948
MH687954
MH687963
MH687964
AYR18468.1
+ More
AYR18482.1 AYR18514.1 AYR18568.1 AYR18574.1 DQ648858 MH687934 AYR18401.1 MH687946 AYR18474.1 MH687941 AYR18443.1 MH687967 AYR18592.1 KT346372 ANV78631.1 MH687957 AYR18532.1 MH687943 MH687956 AYR18456.1 AYR18526.1 MH687949 AYR18488.1 MH687935 AYR18407.1 MH687962 AYR18562.1 MH687936 MH687959 AYR18413.1 AYR18544.1 MH687938 MH687944 MH687955 MH687958 MH687960 MH687965 AYR18425.1 AYR18462.1 AYR18520.1 AYR18538.1 AYR18550.1 AYR18580.1 MH687939 AYR18431.1 MH687950 AYR18494.1 MH687937 MH687961 MH687966 AYR18419.1 AYR18556.1 AYR18586.1 MH687951 AYR18500.1 MH687952 AYR18506.1 MH687940 AYR18437.1 MH687942 AYR18449.1 MN065811 QER90705.1 MG923574 QDE55566.1 GU372739 ADC67068.1 GU372735 GU372741 MH006960 MH006962 MH006963 ADC67064.1 ADC67070.1 AVU05432.1 AVU05446.1 AVU05453.1 KT357514 KT357515 KT357516 KT357518 KT357519 KT357520 ALR34682.1 ALR34683.1 ALR34684.1 ALR34686.1 ALR34687.1 ALR34688.1 KM890281 AJF94315.1 MH744183 QBF53852.1 MH744223 QBF53892.1 MH909809 QCE31487.1 MH744191 QBF53860.1 MH744190 QBF53859.1 KX791060 AQX36343.1 MH744171 QBF53840.1 MH744170 QBF53839.1 MH744173 QBF53842.1 MH744172 QBF53841.1 MH744218 QBF53887.1 MH744244 QBF53913.1 MH744162 QBF53831.1 MH744248 QBF53917.1 MH744184 QBF53853.1 MH744245 QBF53914.1 MH744246 QBF53915.1 MH744214 QBF53883.1 MH744181 QBF53850.1 MH744237 QBF53906.1 MH744163 QBF53832.1 MH744236 QBF53905.1 MH744205 QBF53874.1 MH744220 QBF53889.1 MH744219 QBF53888.1 MH744247 QBF53916.1 MH744158 QBF53827.1 MH744188 QBF53857.1 MH744212 QBF53881.1 MH744189 QBF53858.1 MH744195 QBF53864.1 MH744194 QBF53863.1 MH744200 QBF53869.1 MH744201 QBF53870.1 MH744192 QBF53861.1 MH744196 QBF53865.1 MH744193 QBF53862.1 MH744209 QBF53878.1 MH744230 QBF53899.1 KP222525 AKC35352.1 KJ857447 AJD09570.1 KJ196348 KX580953 AHW40031.1 AQK38169.1 JX501329 AFV59251.1 JX880081 AGA16505.1 MF611899 MF611908 MF611909 AWX65670.1 AWX65679.1 AWX65680.1 KP281650 KP281651 AKA64557.1 AKA64558.1 KJ857441 AJD09564.1 KU641675 ANB44410.1 MG546689 AYP73640.1 JQ664302 KX852322 KX852331 KX852332 KX852333 KX852334 KX852335 AFK09785.1 ASK39728.1 ASK39737.1 ASK39738.1 ASK39739.1 ASK39740.1 ASK39741.1 KU641667 ANB44402.1 KC120788 KP222532 AGD93110.1 AKC35359.1 KF272920 KJ645647 KJ645661 KJ645662 KJ645666 KJ645673 KJ645674 KJ645677 KJ645678 KJ645679 KJ645686 KJ645688 KR265771 KR265787 KR265794 KR265798 KR265804 KR265814 KR265816 KR265818 KU982974 MF373643 AGO58925.1 AID56740.1 AID56824.1 AID56830.1 AID56854.1 AID56896.1 AID56902.1 AID56920.1 AID56926.1 AID56932.1 AID56974.1 AID56986.1 AKJ21774.1 AKJ21870.1 AKJ21912.1 AKJ21936.1 AKJ21972.1 AKJ22032.1 AKJ22044.1 AKJ22056.1 ANI85777.1 AUV64574.1 MH744202 QBF53871.1 MK656494 QFG71757.1 MH744203 QBF53872.1 KM048306 AIT56239.1 KU641638 ANB44373.1 KM048298 AIT56231.1 KU641641 ANB44376.1 LT898426 SNQ28011.1 MK606369 QCB65307.1 KT941120 ALR74758.1 KP281643 KP281644 AKA64550.1 AKA64551.1 KT357517 ALR34685.1 MF577027 ASV51732.1 KM890291 KU977498 AJF94323.1 AOZ60340.1 MK250953 AZS27726.1 MH744180 QBF53849.1 KU641649 ANB44384.1 KF495659 KF495663 AHA15401.1 AHA15405.1 KP281645 AKA64552.1 KY928065 ART84251.1
AYR18482.1 AYR18514.1 AYR18568.1 AYR18574.1 DQ648858 MH687934 AYR18401.1 MH687946 AYR18474.1 MH687941 AYR18443.1 MH687967 AYR18592.1 KT346372 ANV78631.1 MH687957 AYR18532.1 MH687943 MH687956 AYR18456.1 AYR18526.1 MH687949 AYR18488.1 MH687935 AYR18407.1 MH687962 AYR18562.1 MH687936 MH687959 AYR18413.1 AYR18544.1 MH687938 MH687944 MH687955 MH687958 MH687960 MH687965 AYR18425.1 AYR18462.1 AYR18520.1 AYR18538.1 AYR18550.1 AYR18580.1 MH687939 AYR18431.1 MH687950 AYR18494.1 MH687937 MH687961 MH687966 AYR18419.1 AYR18556.1 AYR18586.1 MH687951 AYR18500.1 MH687952 AYR18506.1 MH687940 AYR18437.1 MH687942 AYR18449.1 MN065811 QER90705.1 MG923574 QDE55566.1 GU372739 ADC67068.1 GU372735 GU372741 MH006960 MH006962 MH006963 ADC67064.1 ADC67070.1 AVU05432.1 AVU05446.1 AVU05453.1 KT357514 KT357515 KT357516 KT357518 KT357519 KT357520 ALR34682.1 ALR34683.1 ALR34684.1 ALR34686.1 ALR34687.1 ALR34688.1 KM890281 AJF94315.1 MH744183 QBF53852.1 MH744223 QBF53892.1 MH909809 QCE31487.1 MH744191 QBF53860.1 MH744190 QBF53859.1 KX791060 AQX36343.1 MH744171 QBF53840.1 MH744170 QBF53839.1 MH744173 QBF53842.1 MH744172 QBF53841.1 MH744218 QBF53887.1 MH744244 QBF53913.1 MH744162 QBF53831.1 MH744248 QBF53917.1 MH744184 QBF53853.1 MH744245 QBF53914.1 MH744246 QBF53915.1 MH744214 QBF53883.1 MH744181 QBF53850.1 MH744237 QBF53906.1 MH744163 QBF53832.1 MH744236 QBF53905.1 MH744205 QBF53874.1 MH744220 QBF53889.1 MH744219 QBF53888.1 MH744247 QBF53916.1 MH744158 QBF53827.1 MH744188 QBF53857.1 MH744212 QBF53881.1 MH744189 QBF53858.1 MH744195 QBF53864.1 MH744194 QBF53863.1 MH744200 QBF53869.1 MH744201 QBF53870.1 MH744192 QBF53861.1 MH744196 QBF53865.1 MH744193 QBF53862.1 MH744209 QBF53878.1 MH744230 QBF53899.1 KP222525 AKC35352.1 KJ857447 AJD09570.1 KJ196348 KX580953 AHW40031.1 AQK38169.1 JX501329 AFV59251.1 JX880081 AGA16505.1 MF611899 MF611908 MF611909 AWX65670.1 AWX65679.1 AWX65680.1 KP281650 KP281651 AKA64557.1 AKA64558.1 KJ857441 AJD09564.1 KU641675 ANB44410.1 MG546689 AYP73640.1 JQ664302 KX852322 KX852331 KX852332 KX852333 KX852334 KX852335 AFK09785.1 ASK39728.1 ASK39737.1 ASK39738.1 ASK39739.1 ASK39740.1 ASK39741.1 KU641667 ANB44402.1 KC120788 KP222532 AGD93110.1 AKC35359.1 KF272920 KJ645647 KJ645661 KJ645662 KJ645666 KJ645673 KJ645674 KJ645677 KJ645678 KJ645679 KJ645686 KJ645688 KR265771 KR265787 KR265794 KR265798 KR265804 KR265814 KR265816 KR265818 KU982974 MF373643 AGO58925.1 AID56740.1 AID56824.1 AID56830.1 AID56854.1 AID56896.1 AID56902.1 AID56920.1 AID56926.1 AID56932.1 AID56974.1 AID56986.1 AKJ21774.1 AKJ21870.1 AKJ21912.1 AKJ21936.1 AKJ21972.1 AKJ22032.1 AKJ22044.1 AKJ22056.1 ANI85777.1 AUV64574.1 MH744202 QBF53871.1 MK656494 QFG71757.1 MH744203 QBF53872.1 KM048306 AIT56239.1 KU641638 ANB44373.1 KM048298 AIT56231.1 KU641641 ANB44376.1 LT898426 SNQ28011.1 MK606369 QCB65307.1 KT941120 ALR74758.1 KP281643 KP281644 AKA64550.1 AKA64551.1 KT357517 ALR34685.1 MF577027 ASV51732.1 KM890291 KU977498 AJF94323.1 AOZ60340.1 MK250953 AZS27726.1 MH744180 QBF53849.1 KU641649 ANB44384.1 KF495659 KF495663 AHA15401.1 AHA15405.1 KP281645 AKA64552.1 KY928065 ART84251.1
Proteomes
Pfam
PF03053
Corona_NS3b
Interpro
IPR004293
Coronavirus_Orf3b
ProteinModelPortal
A0A3G3NHP3
Q0Q465
A0A3G3NGB2
A0A3G3NHK5
A0A3G3NHE7
A0A3G3NGZ8
+ More
A0A219TU70 A0A3G3NGT8 A0A3G3NH43 A0A3G3NHL9 A0A3G3NGG5 A0A3G3NGW6 A0A3G3NHB8 A0A3G3NGZ1 A0A3G3NGE6 A0A3G3NHL5 A0A3G3NGF5 A0A3G3NGX3 A0A3G3NH25 A0A3G3NGJ8 A0A3G3NGI5 A0A5J6DDY7 A0A4Y6A773 D3XNW8 D3XNW4 A0A0U2UAH2 A0A0B5INN1 A0A411NPC9 A0A411NPH2 A0A4D6P035 A0A411NPJ9 A0A411NPE4 A0A1S6XZP4 A0A411NPH9 A0A411NPC3 A0A411NPC0 A0A411NPD4 A0A411NPH4 A0A411NPJ0 A0A411NPC1 A0A411NPK3 A0A411NPC8 A0A411NPJ1 A0A411NPK4 A0A411NPG1 A0A411NPI9 A0A411NPL8 A0A411NPB7 A0A411NPJ4 A0A411NPF5 A0A411NPH6 A0A411NPK5 A0A411NPM8 A0A411NPA8 A0A411NPE6 A0A411NPG9 A0A411NPH5 A0A411NPE5 A0A411NPD8 A0A411NPF3 A0A411NPK9 A0A411NPF2 A0A411NPF0 A0A411NPD9 A0A411NPJ5 A0A411NPI6 A0A0E3TJV2 A0A0U1VEK2 X5CC42 K4P5B0 L0CLX1 A0A2Z4N5X0 A0A190LBQ0 A0A0U1YH23 A0A167NFX0 A0A3G3C9B3 I3QJZ9 A0A160EEF0 L7Y082 S0API7 A0A411NPF9 A0A5J6VDZ1 A0A411NPF1 A0A097I514 A0A167NFF4 A0A097I537 A0A160EDB4 A0A238TPN3 A0A4D6FWV9 A0A0U2UEC2 A0A185QEM7 A0A0U2LMD3 A0A286R3Q8 A0A0B5J0Z8 A0A3S9VME3 A0A411NPD3 A0A160EDC3 U6A623 A0A190LBM1 A0A1Y0D9J8
A0A219TU70 A0A3G3NGT8 A0A3G3NH43 A0A3G3NHL9 A0A3G3NGG5 A0A3G3NGW6 A0A3G3NHB8 A0A3G3NGZ1 A0A3G3NGE6 A0A3G3NHL5 A0A3G3NGF5 A0A3G3NGX3 A0A3G3NH25 A0A3G3NGJ8 A0A3G3NGI5 A0A5J6DDY7 A0A4Y6A773 D3XNW8 D3XNW4 A0A0U2UAH2 A0A0B5INN1 A0A411NPC9 A0A411NPH2 A0A4D6P035 A0A411NPJ9 A0A411NPE4 A0A1S6XZP4 A0A411NPH9 A0A411NPC3 A0A411NPC0 A0A411NPD4 A0A411NPH4 A0A411NPJ0 A0A411NPC1 A0A411NPK3 A0A411NPC8 A0A411NPJ1 A0A411NPK4 A0A411NPG1 A0A411NPI9 A0A411NPL8 A0A411NPB7 A0A411NPJ4 A0A411NPF5 A0A411NPH6 A0A411NPK5 A0A411NPM8 A0A411NPA8 A0A411NPE6 A0A411NPG9 A0A411NPH5 A0A411NPE5 A0A411NPD8 A0A411NPF3 A0A411NPK9 A0A411NPF2 A0A411NPF0 A0A411NPD9 A0A411NPJ5 A0A411NPI6 A0A0E3TJV2 A0A0U1VEK2 X5CC42 K4P5B0 L0CLX1 A0A2Z4N5X0 A0A190LBQ0 A0A0U1YH23 A0A167NFX0 A0A3G3C9B3 I3QJZ9 A0A160EEF0 L7Y082 S0API7 A0A411NPF9 A0A5J6VDZ1 A0A411NPF1 A0A097I514 A0A167NFF4 A0A097I537 A0A160EDB4 A0A238TPN3 A0A4D6FWV9 A0A0U2UEC2 A0A185QEM7 A0A0U2LMD3 A0A286R3Q8 A0A0B5J0Z8 A0A3S9VME3 A0A411NPD3 A0A160EDC3 U6A623 A0A190LBM1 A0A1Y0D9J8
Ontologies
KEGG
Subcellular Location
From MSLVP
Multi-Pass Membrane
From Uniprot
Host membrane
Topology
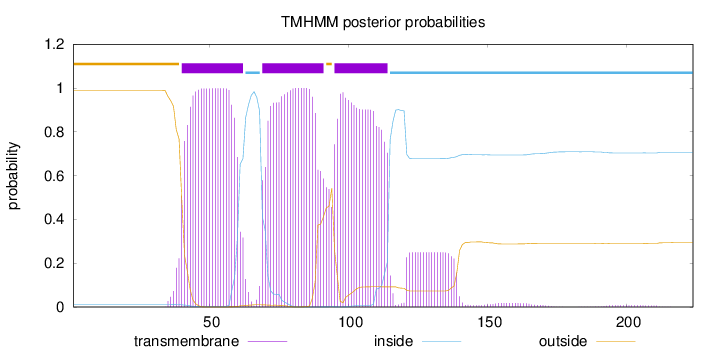
Length:
224
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
65.83469
Exp number, first 60 AAs:
19.93892
Total prob of N-in:
0.01141
POSSIBLE N-term signal
sequence
outside
1 - 39
TMhelix
40 - 62
inside
63 - 68
TMhelix
69 - 91
outside
92 - 94
TMhelix
95 - 114
inside
115 - 224
Population Genetic Test Statistics
Pi
0.3113342
Theta
0.5267128
Tajima's D
-1.5595964
CLR
5.832646
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 |
Protein |
YP_001351685.1 |
|
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
