Strain
CHINA_BAT_2008_NC_010438
(Region: China: Hong Kong; Strain: Bat coronavirus HKU8, complete genome.; Date: 2008)
Gene
spike protein
Description
Annotated in NCBI,
spike protein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGAAATCTTTACTTGTCTTAAGCCTTTTGGCCTTGTTGGCCACATTGTCTGTCAATGCGCAGGTTACTGGTGAAGGCGGTGTGGCAGAAGGTAATTATTGGGTTAATGCCTCTTGTGCTGGTTGGAGCTATTTTTATGCTCTTAAGCTTGGCCTACCACCCAATGCCTCTGCCATAGTCACTGGTTATTTACCCAAACCTAAGGGTTGGATATGTCCTAGATTTAATGGTGCCGGTATCTATACACGTAATAATGCCAATGCTGTTTTTGTTATGTATAGAACCAAAGCCCTTGCCTTTGAGATTGGTGTTAGTTCTTCTGCTGGTGGTGAGCAGTATAGTGCTTACATGGCTCAGCAGAATGGCAAATACCTTGTGCTTCGTATATGCAAGTGGCAGAATGGCACTTTGGCTGCCCCCACTTTGCAGTCCACTTCTGGGAAGGATTGTATTGTCAATGTTAAAGTTGACAATCATATGTTCTATCATGCAGCTCATGATATAGTCGGCATGTCCTGGTCTGGTGACGCTGTACGTCTATATACCCAGACTGACACAAAAACTTATTACATCCCTAATTCTTGGGACAGAGTGTCTATTAGGTGCCCTGATAAGTTCTCGTGTTCCTCACAAATTGTCACTAAGGCTATTACTGTCAATGTCACCACTTTTGCTAATGGTACCATTGACAAGTATGCCATTTGTGACAATTGTAATGGATATCCGGCTCACATCTTTCCTGTATCTGAAGGTGGACTTATTCCAGCTGATTTTAATTTTTCTAACTGGTTCCTGCTTACTAATAGTTCTACTATTGTTGATGGCCGCATTGTTTCAGAACAGCCTGTGCTACTTATGTGTCTTTGGGCTGTACCTGGACTGATGTCAACTAATAGCTTTGTCTATTTCAATGGTACAGCCCCTAATAAGCAATGCAATGGCTATGCTACGGATAGTGCTTTTGAGGCATTACGGTTTTCCCTTAATTTTACCGATGAAAGAGTCTTTGCTGGCAGCGGCAGTGTTGTACTACTTGTGTCTGGTTTGCAGTACAAGTTTAGTTGCACTAATAATTCTGAGGCTGTGATCGATTCCGGAATTCCTTTTGGTAATGTTGTTGAGCCGTTTTATTGTTTTGTCTCTATTAATGGCACTTCTATTTTTGTTGGCATGCTCCCTGCTGTTTTACGTGAAATTGTTATCACTCGTTATGGCAGTATTTATCTCAATGGTTTCTCCATTTTTCAAGGCCCACCCATTCAGGGTGTTTTGTTCAATGTAACTAATAGAGGTGCAACTGATCTTTGGACCGTTGCACTCTCTAACTTTACGGAAGTCCTTGCTGAAGTTCAAAGTACTGCCATTAAAGCTCTGCTTTATTGTGATGACCCATTGTCTCAGCTTAAGTGTCAACAATTGCAATTTTCGCTGCCTGATGGGTTTTATGCAACTGCCTCACTTTTTCAACATGAGTTGCCACGTACTTTTGTCACTTTACCACGCCATTTTACGCATTCTTGGATTAACCTGCGTATTAAATGGAAGAATGGTGTTTGTTACAATTGTCCACCTGCTTCTTCTTGGATTGATTTTGTAACATTCAATTCTAATGGCACCGAGAATGTTCTTCCTGAACGCACTTTATGCGTTAACACCACTCAGTTTACTACCAATTTGACTCTTATTGAGGAGGCTTTTAGTTATTCCACTCCTGTTGTTGTTCGTGCTGATGATTGTCCTTTTGACTTCCAGTCCTTGAACAATTACCTAACTTTTGGTTCCATTTGTTTTTCTTTGAATGGCACCATTGGTAAGGGTTGCACACTTGGAATATATAAACGAGCCTCGTCTCAGTACATTCCTATTTGGAATGTTTGGGTGGCTTACACTAGTGGTGATAATATCCTCGGTGTCAGAGAGCCCAATGTTGGTGTTAGAGATCAGAGTGTAGTGCACCAGAATGTTTGTACATCATATACAATTTTTGGGCATTCAGGTCGCGGTATTATCAGACCTGCTAATATTTCATATATAGCAGGTGTGTATTATACTGCTGCTTCTGGTCAACTATTGGGCTTTAAGAATACCACCACTGGTGAAGTGTTTTCTGTTACACCATGTAACCCATCACAGCAGGCTGTTGTTGTGAAGGATCGTCTTGTTGGTGTTATGTCATCTACTAGCACTGTTTCGATTCCGTTTAATAATACGATTCCAACTCCTAGTTTTTACTATCATAGTAATGCCACTAGTAGTTGTGATGATCCATCTGTTGTTTATTCATCTATTGGTATTTGTGATGATGGCGGTATTACTTTTGTTAATAGTACTAGGGTTCGTGGTGAACCAGATCCTGCTATTTCAATGGGTAATATTTCTGTTCCATCAAATTTCACTGTTTCCATTCAGGTTGAGTACTTGCAGATGTCCATTAGGCCTGTTTCGATAGATTGTGCTATGTATGTCTGTAATGGCAATCCTCATTGTACTAGGTTGTTGCAGCAATATATTTCTGCTTGCCGAACCATTGAGGAGGCCTTACAGCTTAGCGCGCGCCTAGAGAGTTTTGAAGTTAATAGCATGCTGACTGTTTCTGAAACAGCGTTAGACCTAGTCAACATCAGCACATTTGGTGGCGATTACAATCTCACTGCTTTGTTGCCGCAAGGTGGTGGTAAACGCAGTGTTATTGAAGATATCTTGTTTGACAAGGTTGTCACCAGTGGGCTGGGCACTGTTGATGAGGATTACAAGCGTTGTACCAATGGTATTGGTATTGCTGATGTCCCATGTGCGCAGTATTATAATGGTATTATGGTACTGCCAGGTGTTGTTGATGAGGAGAAGATGTCCATCTATACTGCTTCCCTTCTTGGTGGCATGACTATGGGTGGTTTTACACCTGTCGCTGCTTTGCCTTTTGCTTTGTCGGTACAGTCCCGTCTTAATTATGTTGCACTCCAAACTGATGTTTTGCAGAAAAATCAACAGATTTTGGCAAATGCCTTTAATTCTGCAATTGGTAATATCACTGTTGCGTTTGATCAGGTAACAACTGCAGTGCAACAAACTTCTGATGCTATTAAAACTGTTGCCAGCGCTCTTAATAAGGTTCAGAGCGTTGTTAACTCTCAAGGTCAGGCCTTGCATCAACTTACTAAACAGCTTGCTTCCAATTTTCAAGCTATTTCTGCTTCTATTGAAGATATTTATAATAGGCTTGATGGGCTTGCTGCTGATGCTAATGTTGATCGCTTAATTACAGGACGCCTTGCTGCTTTGAATGCTTTTGTTACTCAGACTCTTACTAAGTACACAGAAGTTCGTGCTAGCAGGTTGTTGGCGCAGGAGAAGATTAATGAGTGTGTTAAGTCACAATCCACTCGTTATGGATTTTGTGGAAATGGCACTCATTTGTTCTCCATCCCTAATGCTGCACCTGAAGGTATTATGCTGTTTCATACAGTTCTTGTACCCACTGAATATGTCTCAGTTACTGCCTGGTCTGGCTATTGTCATAACGGTGTTGGTTATGCCGTGAAAGATGTAGGCAATTCGCTGTTTCAGTTTAATAATACATTTTACATCACACCTCGAAATATGTATCAGCCACGTACACCTACGTCCGCTGACTTTATTCGTATTTCAGGTTGTAATGTTGTCTATGTCAACATTACTGATGAACAGCTGCCACAAGTTCAGCCTGAGTTTATTGATGTCAACAAGACTCTTGAAGAGTTAATGTCCAGGTTGCCAAACAATACAGGGCCGAACTTGCCTCTTGATATTTTTAACCAGACATATCTTAATATCAGTGCTGAGATTGATGCTCTTGAAAATAAGTCATTGGAGCTTCAGGCTACTGCTGACAAATTACAGCTAACCATTGAGCAGCTCAATGCCACTCTCGTTGATCTTGAATGGCTTAATCGCTTTGAACAGTATGTTAAGTGGCCCTGGTGGGTATGGCTCACCATGATTATTGCCTTGGTCTTGTTGACTGGGCTTATGTTATGGTGTTGCCTTGCTACTGGATGCTGTGGTTGCTGTAGTTGTATGGCTAGTACTCTTGATTTTAGAGGCAGTAGGTTACAACAATACGAAGTTGAAAAAGTGCACATCCAGTAA
Protein
MKSLLVLSLLALLATLSVNAQVTGEGGVAEGNYWVNASCAGWSYFYALKLGLPPNASAIVTGYLPKPKGWICPRFNGAGIYTRNNANAVFVMYRTKALAFEIGVSSSAGGEQYSAYMAQQNGKYLVLRICKWQNGTLAAPTLQSTSGKDCIVNVKVDNHMFYHAAHDIVGMSWSGDAVRLYTQTDTKTYYIPNSWDRVSIRCPDKFSCSSQIVTKAITVNVTTFANGTIDKYAICDNCNGYPAHIFPVSEGGLIPADFNFSNWFLLTNSSTIVDGRIVSEQPVLLMCLWAVPGLMSTNSFVYFNGTAPNKQCNGYATDSAFEALRFSLNFTDERVFAGSGSVVLLVSGLQYKFSCTNNSEAVIDSGIPFGNVVEPFYCFVSINGTSIFVGMLPAVLREIVITRYGSIYLNGFSIFQGPPIQGVLFNVTNRGATDLWTVALSNFTEVLAEVQSTAIKALLYCDDPLSQLKCQQLQFSLPDGFYATASLFQHELPRTFVTLPRHFTHSWINLRIKWKNGVCYNCPPASSWIDFVTFNSNGTENVLPERTLCVNTTQFTTNLTLIEEAFSYSTPVVVRADDCPFDFQSLNNYLTFGSICFSLNGTIGKGCTLGIYKRASSQYIPIWNVWVAYTSGDNILGVREPNVGVRDQSVVHQNVCTSYTIFGHSGRGIIRPANISYIAGVYYTAASGQLLGFKNTTTGEVFSVTPCNPSQQAVVVKDRLVGVMSSTSTVSIPFNNTIPTPSFYYHSNATSSCDDPSVVYSSIGICDDGGITFVNSTRVRGEPDPAISMGNISVPSNFTVSIQVEYLQMSIRPVSIDCAMYVCNGNPHCTRLLQQYISACRTIEEALQLSARLESFEVNSMLTVSETALDLVNISTFGGDYNLTALLPQGGGKRSVIEDILFDKVVTSGLGTVDEDYKRCTNGIGIADVPCAQYYNGIMVLPGVVDEEKMSIYTASLLGGMTMGGFTPVAALPFALSVQSRLNYVALQTDVLQKNQQILANAFNSAIGNITVAFDQVTTAVQQTSDAIKTVASALNKVQSVVNSQGQALHQLTKQLASNFQAISASIEDIYNRLDGLAADANVDRLITGRLAALNAFVTQTLTKYTEVRASRLLAQEKINECVKSQSTRYGFCGNGTHLFSIPNAAPEGIMLFHTVLVPTEYVSVTAWSGYCHNGVGYAVKDVGNSLFQFNNTFYITPRNMYQPRTPTSADFIRISGCNVVYVNITDEQLPQVQPEFIDVNKTLEELMSRLPNNTGPNLPLDIFNQTYLNISAEIDALENKSLELQATADKLQLTIEQLNATLVDLEWLNRFEQYVKWPWWVWLTMIIALVLLTGLMLWCCLATGCCGCCSCMASTLDFRGSRLQQYEVEKVHIQ
Summary
Function
S1 region attaches the virion to the cell membrane by interacting with host ANPEP/aminopeptidase N, initiating the infection. Binding to the receptor probably induces conformational changes in the S glycoprotein unmasking the fusion peptide of S2 region and activating membranes fusion. S2 region belongs to the class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) regions assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer. During virus morphogenesis, found in a complex with M and HE proteins. Interacts with host ANPEP.
Similarity
Belongs to the alphacoronaviruses spike protein family.
Uniprot
B1PHK2
A0A0U1WHD0
F1DB20
A0A1L2KGD0
A0A482JX42
A0A482JZV2
+ More
A0A0U1WJW2 A4ULL1 K4JZ69 K4JZP8 K4K2R0 A0A0U1WHD9 A0A0U1UZ37 A0A4Y5QL66 A0A0U1UYV9 K4JZD2 K4K211 A0A482K0W7 A0A4Y6GL65 F1DAX6 A0A482JZK4 A0A0U1WHB6 A0A1W5YKS6 A0A0U1WHD7 A0A1W5YKT3 F1DB14 K4JZU5 K4KCP3 A0A0U1WHE4 F1DB07 A0A126K9I9 A0A1L2KGB2 A0A126K9A8 A0A1Z1W238 A0A1L2KGD3 A0A0U1WHB3 A0A0U1UZ27 A0A126KBA9 A0A126K9U6 A0A126K9J8 B1PHJ5 A0A0U1WHB7 A0A126KBA2 A0A1Z1W245 A0A126KB98 A0A1Z1W240 A0A1Z1W228 A0A1Z1W250 A0A126KBA0 A0A0U1WHD6 A0A1Z1W243 A0A1Z1W231 A0A1L2KGF1 B1PHI8 A0A0N7J129 A0A3G8EWN4 A0A4Y5QL97 A0A0P0KI80 A0A0P0KR98 A0A0P0K0X5 A0A0N7J128 A0A0P0KXK6 A0A0P0KBC5 A0A1L2KGE4 A0A3G8EWC1 A0A3G8EWD5 A0A0P0KMM1 A0A0P0KR90 A0A0P0K6L9 A4ULL0 A0A0U1UZD0 A0A220YLM0 A0A0P0KRB3 A0A0P0KWD4 A0A0P0KH07 A0A286R3Q7 S5YNL4 A0A3G3NGQ9 A0A3G3NGQ6 A0A3G3NGX2 A0A3G3NGH9 A0A3G3NGU0 A0A3G3NH26 A0A3G3NGU7 A0A3G3NH85 A0A3G3NHI1 A0A3G3NH56 A0A1L3GVA2 A0A023ULP1 A0A0U2L0Z5 A0A3G3NHJ6 A0A3S8TLM6 A0A0U1YG53 A0A516KAN2 A0A222NUN9 A0A5A4M032 D1MJB7 I7BQC9 A0A1L3KJ30 A0A0U1YG21
A0A0U1WJW2 A4ULL1 K4JZ69 K4JZP8 K4K2R0 A0A0U1WHD9 A0A0U1UZ37 A0A4Y5QL66 A0A0U1UYV9 K4JZD2 K4K211 A0A482K0W7 A0A4Y6GL65 F1DAX6 A0A482JZK4 A0A0U1WHB6 A0A1W5YKS6 A0A0U1WHD7 A0A1W5YKT3 F1DB14 K4JZU5 K4KCP3 A0A0U1WHE4 F1DB07 A0A126K9I9 A0A1L2KGB2 A0A126K9A8 A0A1Z1W238 A0A1L2KGD3 A0A0U1WHB3 A0A0U1UZ27 A0A126KBA9 A0A126K9U6 A0A126K9J8 B1PHJ5 A0A0U1WHB7 A0A126KBA2 A0A1Z1W245 A0A126KB98 A0A1Z1W240 A0A1Z1W228 A0A1Z1W250 A0A126KBA0 A0A0U1WHD6 A0A1Z1W243 A0A1Z1W231 A0A1L2KGF1 B1PHI8 A0A0N7J129 A0A3G8EWN4 A0A4Y5QL97 A0A0P0KI80 A0A0P0KR98 A0A0P0K0X5 A0A0N7J128 A0A0P0KXK6 A0A0P0KBC5 A0A1L2KGE4 A0A3G8EWC1 A0A3G8EWD5 A0A0P0KMM1 A0A0P0KR90 A0A0P0K6L9 A4ULL0 A0A0U1UZD0 A0A220YLM0 A0A0P0KRB3 A0A0P0KWD4 A0A0P0KH07 A0A286R3Q7 S5YNL4 A0A3G3NGQ9 A0A3G3NGQ6 A0A3G3NGX2 A0A3G3NGH9 A0A3G3NGU0 A0A3G3NH26 A0A3G3NGU7 A0A3G3NH85 A0A3G3NHI1 A0A3G3NH56 A0A1L3GVA2 A0A023ULP1 A0A0U2L0Z5 A0A3G3NHJ6 A0A3S8TLM6 A0A0U1YG53 A0A516KAN2 A0A222NUN9 A0A5A4M032 D1MJB7 I7BQC9 A0A1L3KJ30 A0A0U1YG21
Pubmed
EMBL
EU420139
ACA52171.1
KJ473798
AIA62220.1
HQ728486
ADX59495.1
+ More
KY073746 APD51491.1 MG916902 QBP43268.1 MG916903 QBP43279.1 KJ473807 AIA62252.1 EF434381 ABO88151.1 JQ989268 AFU92086.1 JQ989266 JQ989267 AFU92070.1 AFU92078.1 JQ989269 AFU92095.1 KJ473800 AIA62234.1 KJ473810 AIA62271.1 MK720945 MK720946 QCX35167.1 QCX35178.1 KJ473799 AIA62227.1 JQ989273 AFU92131.1 JQ989272 AFU92122.1 MG916904 QBP43290.1 MK211373 QDF43810.1 HQ728480 ADX59451.1 MG916901 QBP43256.1 KJ473797 AIA62212.1 KY770850 ARI44789.1 KJ473806 AIA62246.1 KY770851 ARI44794.1 HQ728485 ADX59488.1 JQ989271 AFU92113.1 JQ989270 AFU92104.1 KJ473803 AIA62242.1 HQ728484 ADX59482.1 KU343205 AMB43195.1 KY073744 APD51475.1 KU343208 AMB43198.1 KY783847 ARX80143.1 KY073745 APD51483.1 KJ473795 AIA62200.1 KJ473802 AIA62241.1 KU343206 AMB43196.1 KU343202 AMB43192.1 KU343207 AMB43197.1 EU420138 ACA52164.1 KJ473796 AIA62206.1 KU343204 AMB43194.1 KY783850 ARX80146.1 KU343203 AMB43193.1 KY783848 ARX80144.1 KY783845 ARX80141.1 KY783851 ARX80147.1 KU343201 AMB43191.1 KJ473801 AIA62240.1 KY783849 ARX80145.1 KY783846 ARX80142.1 KY073748 APD51507.1 EU420137 ACA52157.1 KT253269 ALK28775.1 MH938450 AZF86130.1 MK720944 QCX35160.1 KT253266 ALK28772.1 KT253270 ALK28781.1 KT253265 ALK28771.1 KT253261 KT253263 ALK28767.1 ALK28769.1 KT253262 ALK28768.1 KT253267 ALK28773.1 KY073747 APD51499.1 MH938448 AZF86118.1 MH938449 AZF86124.1 KT253268 ALK28774.1 KT253264 ALK28770.1 KT253271 ALK28787.1 EF434379 ABO88150.1 KJ473809 AIA62265.1 KY799179 ASL24654.1 KT253272 ALK28793.1 KT253259 ALK28765.1 KT253260 ALK28766.1 MF577027 ASV51733.1 KF430219 AGT21333.1 MH687955 AYR18519.1 MH687957 AYR18531.1 MH687965 AYR18579.1 MH687944 AYR18461.1 MH687960 AYR18549.1 MH687956 AYR18525.1 MH687958 AYR18537.1 MH687966 AYR18585.1 MH687937 AYR18418.1 MH687961 AYR18555.1 KY019624 APG31005.1 KJ451037 AHY04180.1 KR061459 ALR84915.1 MH687942 AYR18448.1 MH593141 AZL47242.1 KJ857476 AJD09599.1 MK410092 QDP38456.1 KX982564 ASQ43221.1 MG198634 AXS76458.1 GU180148 ACZ52971.1 JX018180 AFO42861.1 KX883635 APG77336.1 KJ857474 AJD09597.1
KY073746 APD51491.1 MG916902 QBP43268.1 MG916903 QBP43279.1 KJ473807 AIA62252.1 EF434381 ABO88151.1 JQ989268 AFU92086.1 JQ989266 JQ989267 AFU92070.1 AFU92078.1 JQ989269 AFU92095.1 KJ473800 AIA62234.1 KJ473810 AIA62271.1 MK720945 MK720946 QCX35167.1 QCX35178.1 KJ473799 AIA62227.1 JQ989273 AFU92131.1 JQ989272 AFU92122.1 MG916904 QBP43290.1 MK211373 QDF43810.1 HQ728480 ADX59451.1 MG916901 QBP43256.1 KJ473797 AIA62212.1 KY770850 ARI44789.1 KJ473806 AIA62246.1 KY770851 ARI44794.1 HQ728485 ADX59488.1 JQ989271 AFU92113.1 JQ989270 AFU92104.1 KJ473803 AIA62242.1 HQ728484 ADX59482.1 KU343205 AMB43195.1 KY073744 APD51475.1 KU343208 AMB43198.1 KY783847 ARX80143.1 KY073745 APD51483.1 KJ473795 AIA62200.1 KJ473802 AIA62241.1 KU343206 AMB43196.1 KU343202 AMB43192.1 KU343207 AMB43197.1 EU420138 ACA52164.1 KJ473796 AIA62206.1 KU343204 AMB43194.1 KY783850 ARX80146.1 KU343203 AMB43193.1 KY783848 ARX80144.1 KY783845 ARX80141.1 KY783851 ARX80147.1 KU343201 AMB43191.1 KJ473801 AIA62240.1 KY783849 ARX80145.1 KY783846 ARX80142.1 KY073748 APD51507.1 EU420137 ACA52157.1 KT253269 ALK28775.1 MH938450 AZF86130.1 MK720944 QCX35160.1 KT253266 ALK28772.1 KT253270 ALK28781.1 KT253265 ALK28771.1 KT253261 KT253263 ALK28767.1 ALK28769.1 KT253262 ALK28768.1 KT253267 ALK28773.1 KY073747 APD51499.1 MH938448 AZF86118.1 MH938449 AZF86124.1 KT253268 ALK28774.1 KT253264 ALK28770.1 KT253271 ALK28787.1 EF434379 ABO88150.1 KJ473809 AIA62265.1 KY799179 ASL24654.1 KT253272 ALK28793.1 KT253259 ALK28765.1 KT253260 ALK28766.1 MF577027 ASV51733.1 KF430219 AGT21333.1 MH687955 AYR18519.1 MH687957 AYR18531.1 MH687965 AYR18579.1 MH687944 AYR18461.1 MH687960 AYR18549.1 MH687956 AYR18525.1 MH687958 AYR18537.1 MH687966 AYR18585.1 MH687937 AYR18418.1 MH687961 AYR18555.1 KY019624 APG31005.1 KJ451037 AHY04180.1 KR061459 ALR84915.1 MH687942 AYR18448.1 MH593141 AZL47242.1 KJ857476 AJD09599.1 MK410092 QDP38456.1 KX982564 ASQ43221.1 MG198634 AXS76458.1 GU180148 ACZ52971.1 JX018180 AFO42861.1 KX883635 APG77336.1 KJ857474 AJD09597.1
Proteomes
UP000143524
UP000110898
UP000106394
UP000123997
UP000126615
UP000132096
+ More
UP000119818 UP000126502 UP000144177 UP000135768 UP000126962 UP000112496 UP000106776 UP000144778 UP000162430 UP000155419 UP000204102 UP000277178 UP000137012 UP000100808 UP000157060 UP000147155 UP000118770 UP000122373 UP000096398 UP000103794 UP000130985 UP000174423 UP000125818
UP000119818 UP000126502 UP000144177 UP000135768 UP000126962 UP000112496 UP000106776 UP000144778 UP000162430 UP000155419 UP000204102 UP000277178 UP000137012 UP000100808 UP000157060 UP000147155 UP000118770 UP000122373 UP000096398 UP000103794 UP000130985 UP000174423 UP000125818
ProteinModelPortal
B1PHK2
A0A0U1WHD0
F1DB20
A0A1L2KGD0
A0A482JX42
A0A482JZV2
+ More
A0A0U1WJW2 A4ULL1 K4JZ69 K4JZP8 K4K2R0 A0A0U1WHD9 A0A0U1UZ37 A0A4Y5QL66 A0A0U1UYV9 K4JZD2 K4K211 A0A482K0W7 A0A4Y6GL65 F1DAX6 A0A482JZK4 A0A0U1WHB6 A0A1W5YKS6 A0A0U1WHD7 A0A1W5YKT3 F1DB14 K4JZU5 K4KCP3 A0A0U1WHE4 F1DB07 A0A126K9I9 A0A1L2KGB2 A0A126K9A8 A0A1Z1W238 A0A1L2KGD3 A0A0U1WHB3 A0A0U1UZ27 A0A126KBA9 A0A126K9U6 A0A126K9J8 B1PHJ5 A0A0U1WHB7 A0A126KBA2 A0A1Z1W245 A0A126KB98 A0A1Z1W240 A0A1Z1W228 A0A1Z1W250 A0A126KBA0 A0A0U1WHD6 A0A1Z1W243 A0A1Z1W231 A0A1L2KGF1 B1PHI8 A0A0N7J129 A0A3G8EWN4 A0A4Y5QL97 A0A0P0KI80 A0A0P0KR98 A0A0P0K0X5 A0A0N7J128 A0A0P0KXK6 A0A0P0KBC5 A0A1L2KGE4 A0A3G8EWC1 A0A3G8EWD5 A0A0P0KMM1 A0A0P0KR90 A0A0P0K6L9 A4ULL0 A0A0U1UZD0 A0A220YLM0 A0A0P0KRB3 A0A0P0KWD4 A0A0P0KH07 A0A286R3Q7 S5YNL4 A0A3G3NGQ9 A0A3G3NGQ6 A0A3G3NGX2 A0A3G3NGH9 A0A3G3NGU0 A0A3G3NH26 A0A3G3NGU7 A0A3G3NH85 A0A3G3NHI1 A0A3G3NH56 A0A1L3GVA2 A0A023ULP1 A0A0U2L0Z5 A0A3G3NHJ6 A0A3S8TLM6 A0A0U1YG53 A0A516KAN2 A0A222NUN9 A0A5A4M032 D1MJB7 I7BQC9 A0A1L3KJ30 A0A0U1YG21
A0A0U1WJW2 A4ULL1 K4JZ69 K4JZP8 K4K2R0 A0A0U1WHD9 A0A0U1UZ37 A0A4Y5QL66 A0A0U1UYV9 K4JZD2 K4K211 A0A482K0W7 A0A4Y6GL65 F1DAX6 A0A482JZK4 A0A0U1WHB6 A0A1W5YKS6 A0A0U1WHD7 A0A1W5YKT3 F1DB14 K4JZU5 K4KCP3 A0A0U1WHE4 F1DB07 A0A126K9I9 A0A1L2KGB2 A0A126K9A8 A0A1Z1W238 A0A1L2KGD3 A0A0U1WHB3 A0A0U1UZ27 A0A126KBA9 A0A126K9U6 A0A126K9J8 B1PHJ5 A0A0U1WHB7 A0A126KBA2 A0A1Z1W245 A0A126KB98 A0A1Z1W240 A0A1Z1W228 A0A1Z1W250 A0A126KBA0 A0A0U1WHD6 A0A1Z1W243 A0A1Z1W231 A0A1L2KGF1 B1PHI8 A0A0N7J129 A0A3G8EWN4 A0A4Y5QL97 A0A0P0KI80 A0A0P0KR98 A0A0P0K0X5 A0A0N7J128 A0A0P0KXK6 A0A0P0KBC5 A0A1L2KGE4 A0A3G8EWC1 A0A3G8EWD5 A0A0P0KMM1 A0A0P0KR90 A0A0P0K6L9 A4ULL0 A0A0U1UZD0 A0A220YLM0 A0A0P0KRB3 A0A0P0KWD4 A0A0P0KH07 A0A286R3Q7 S5YNL4 A0A3G3NGQ9 A0A3G3NGQ6 A0A3G3NGX2 A0A3G3NGH9 A0A3G3NGU0 A0A3G3NH26 A0A3G3NGU7 A0A3G3NH85 A0A3G3NHI1 A0A3G3NH56 A0A1L3GVA2 A0A023ULP1 A0A0U2L0Z5 A0A3G3NHJ6 A0A3S8TLM6 A0A0U1YG53 A0A516KAN2 A0A222NUN9 A0A5A4M032 D1MJB7 I7BQC9 A0A1L3KJ30 A0A0U1YG21
PDB
6U7K
E-value=0,
Score=2847
Ontologies
KEGG
GO
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0016021 C:integral component of membrane
GO:0019031 C:viral envelope
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0016021 C:integral component of membrane
GO:0019031 C:viral envelope
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0055036 C:virion membrane
GO:0009405 P:pathogenesis
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0039654 P:fusion of virus membrane with host endosome membrane
Subcellular Location
From MSLVP
Single-Pass Membrane
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
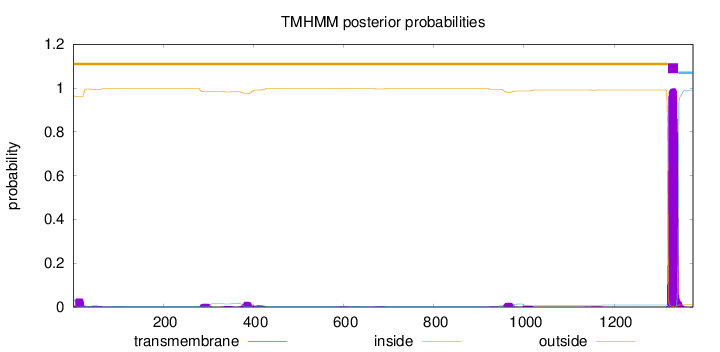
Topology
Length:
1375
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.80815
Exp number, first 60 AAs:
0.88041
Total prob of N-in:
0.03664
outside
1 - 1319
TMhelix
1320 - 1342
inside
1343 - 1375
Population Genetic Test Statistics
Pi
0.3285906
Theta
0.559143
Tajima's D
-1.619729
CLR
9.497041
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 |
Protein |
YP_001718612.1 |
|
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.