Strain
AMERICA_WHALE_2007_NC_010646
(Region: North America; Strain: Beluga Whale coronavirus SW1, complete genome.; Date: 2007)
Gene
matrix glycoprotein
Description
Annotated in NCBI,
matrix glycoprotein
Location
GenBank Accession
Full name
Membrane protein
Alternative Name
E1 glycoprotein
Matrix glycoprotein
Membrane glycoprotein
Matrix glycoprotein
Membrane glycoprotein
Sequence
CDS
ATGGTGAATATGTATGAAGTAGCGCTTCGTGTGCTTCGTGATTACAATTTAGCATTATCAGCGTTTCTGACATTGATTATTTGTGTGTTACAGTTTGGTTATGCCTCCCGTAACAGGTTCTTTTATTGGATTAAGATTATTACTTTGTGGCTTATGTGGCCGTTTGCTATTGTAACGTCGGTGTTTTCATGTTTGTATCCACCATGTTATAGTTATGCCTCTAGGTTTAGTAATAACACTTCCACCACTCCAACACCAGTTGAAGGTAATGGTTTGTGGTCATGTGGTTCTAGTTATGCAATTTCTGTTAACCTTACTGCATTTGTGTTTGCAATAATTTTTGCAATTTTCACATGTGCGGCATGGCTATTTTATTGGATACAATCCACACGCCTGTATTTGAGAGTAAGATCATGGTGGGCTTTCTCACCAGACACTAACTACGTTGCTGTATTTAAAATGGCTAATGACCAGTATTTCACGCTGCCTTTGCTTGAAGTGCCTATGGTTCTTAGTCTTGTTTATAAGAATAATTTTGTTTATTGTAATGGTACATATCTTGGCAGAATACCCAGTATGGGTTATTTACCCGCTAGTTGTATAGTAGGTAGTGCCGAGAAAATTGCTAGATACAGAAAAATTACAGTTTCAGCAGCAGATGGCAGTGATGCTAATGTTGCGCGTTTTGGTACTGTGATTTATGCTAAAGATAAATATATAAGTGCAGGGCTTGCGTATACTAAGAGAGAGAAAACTGAGACAGAGAAGCTTTATTCCGTTTAA
Protein
MVNMYEVALRVLRDYNLALSAFLTLIICVLQFGYASRNRFFYWIKIITLWLMWPFAIVTSVFSCLYPPCYSYASRFSNNTSTTPTPVEGNGLWSCGSSYAISVNLTAFVFAIIFAIFTCAAWLFYWIQSTRLYLRVRSWWAFSPDTNYVAVFKMANDQYFTLPLLEVPMVLSLVYKNNFVYCNGTYLGRIPSMGYLPASCIVGSAEKIARYRKITVSAADGSDANVARFGTVIYAKDKYISAGLAYTKREKTETEKLYSV
Summary
Function
Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins.
Subunit
Homomultimer. Interacts with envelope E protein in the budding compartment of the host cell, which is located between endoplasmic reticulum and the Golgi complex. Forms a complex with HE and S proteins. Interacts with nucleocapsid N protein. This interaction probably participates in RNA packaging into the virus.
Similarity
Belongs to the gammacoronaviruses M protein family.
Keywords
Glycoprotein
Host Golgi apparatus
Host membrane
Membrane
Transmembrane
Transmembrane helix
Viral envelope protein
Viral matrix protein
Virion
Reference proteome
Feature
chain Membrane protein
Uniprot
B2BW35
V5TGF3
V5TFF2
C5IZI4
B3VZS5
A0A481NU89
+ More
C5IZJ0 C5IZJ2 C5IZI5 C5IZI6 A0A5J6RCQ6 G4RJ98 G3C785 K7RWV4 C5IUA3 C6KCX8 A5Z0Z4 A0A5J6RC67 A0A1W6AWK4 K7S5A6 C1KAC3 C6KCX1 A0A140HEW9 A0A0A1G0F9 A0A5J6RDV5 E9LUF4 A4ZCQ8 A5Z142 C6KCY2 B3GPG8 A0A5J6RDK2 A0A5J6RDL5 J9WMJ2 J9WRY4 A0A1B2CWH4 A0A0U1WJZ5 G4RJ75 A0A1B0Z544 P12649 A0A1P8SFM3 A0A1P8FCL8 C5IUA4 Q6DTU8 Q64FZ0 A5Z149 C6KCX4 C6KCX5 A0A140DP47 Q80RY9 A0A1L1XGR3 A0A516UWB7 A0A1P8SMJ8 C6KCY0 U5U9J5 C7DY52 G4RJ95 A0A0D4CG32 C4PE61 F4MIY0 Q9J3N8 J9WLU4 I7B3F0 A0A1W6AWL0 Q6VUR2 P69606 P69607 A0A142IJS1 A0A5J6RDM6 A4ZCM9 Q6R4P1 Q58J43 A4ZCJ2 C7S863 A0A481NUK2 Q7T9P6 G3FEJ2 G4RJ86 A0A1P8FCE8 C9DST0 B3VN11 C7S7Z4 U5YDU7 Q7TEH1 A0A0C5GXD2 Q91SA2 A0A4V6MBA0 Q6W9D6 P69604 P69603 P69605 I7AZ79 A0A346M350 G4RJA1 A4ZCV6 A0A1P8VF79 A0A0U1UZF0 A0A0U1WK58 Q910H8 A0A346M361
C5IZJ0 C5IZJ2 C5IZI5 C5IZI6 A0A5J6RCQ6 G4RJ98 G3C785 K7RWV4 C5IUA3 C6KCX8 A5Z0Z4 A0A5J6RC67 A0A1W6AWK4 K7S5A6 C1KAC3 C6KCX1 A0A140HEW9 A0A0A1G0F9 A0A5J6RDV5 E9LUF4 A4ZCQ8 A5Z142 C6KCY2 B3GPG8 A0A5J6RDK2 A0A5J6RDL5 J9WMJ2 J9WRY4 A0A1B2CWH4 A0A0U1WJZ5 G4RJ75 A0A1B0Z544 P12649 A0A1P8SFM3 A0A1P8FCL8 C5IUA4 Q6DTU8 Q64FZ0 A5Z149 C6KCX4 C6KCX5 A0A140DP47 Q80RY9 A0A1L1XGR3 A0A516UWB7 A0A1P8SMJ8 C6KCY0 U5U9J5 C7DY52 G4RJ95 A0A0D4CG32 C4PE61 F4MIY0 Q9J3N8 J9WLU4 I7B3F0 A0A1W6AWL0 Q6VUR2 P69606 P69607 A0A142IJS1 A0A5J6RDM6 A4ZCM9 Q6R4P1 Q58J43 A4ZCJ2 C7S863 A0A481NUK2 Q7T9P6 G3FEJ2 G4RJ86 A0A1P8FCE8 C9DST0 B3VN11 C7S7Z4 U5YDU7 Q7TEH1 A0A0C5GXD2 Q91SA2 A0A4V6MBA0 Q6W9D6 P69604 P69603 P69605 I7AZ79 A0A346M350 G4RJA1 A4ZCV6 A0A1P8VF79 A0A0U1UZF0 A0A0U1WK58 Q910H8 A0A346M361
Pubmed
18353961
24227844
31561498
21994806
18295413
23171741
+ More
22966194 18045937 29141204 2841803 27793730 15722532 26482289 26616599 15019237 21539870 11087096 21909766 23203570 28495648 28646651 30526618 16934878 21049275 22542436 25721384 15183070 16927130 23182435 20022075 25843651 19100792 20652731 23178317
22966194 18045937 29141204 2841803 27793730 15722532 26482289 26616599 15019237 21539870 11087096 21909766 23203570 28495648 28646651 30526618 16934878 21049275 22542436 25721384 15183070 16927130 23182435 20022075 25843651 19100792 20652731 23178317
EMBL
EU111742
ABW87822.1
KF793825
AHB63497.1
KF793824
KF793826
+ More
AHB63483.1 AHB63510.1 FJ976459 ACR16060.1 EU826085 ACE95721.1 MH151328 QAV53736.1 FJ976465 ACR16066.1 FJ976467 ACR16068.1 FJ976460 ACR16061.1 FJ976461 ACR16062.1 MK887107 QEY95712.1 FJ849823 ACZ67645.1 GU393332 ADV71743.1 JX565012 MH924835 MK142676 AFV92602.1 AZA07518.1 QCX41899.1 FJ965348 GQ174475 ACR55677.1 ACS45207.1 GQ174481 ACS45213.1 EF602444 ABQ84802.1 MK887111 QEY95716.1 KY626044 ARJ35785.1 JX565011 AFV92601.1 FJ807653 GQ179645 JF893452 KF663560 ACO37578.1 ACS45205.1 AEJ80242.1 AHB74003.1 GQ174474 HQ283475 HQ850618 ACS45206.1 ADT91599.1 AEB00719.1 KT886454 AMO03296.1 KM213963 AIY51832.1 MK887104 MK887108 QEY95709.1 QEY95713.1 HQ141058 ADW77296.1 DQ490214 ABF61535.1 EF602450 KY799582 ABQ84850.1 AVI69489.1 GQ174485 ACS45217.1 EU714034 GQ227602 KF377577 KP036503 KP036504 KP118880 KP118881 KP118882 KP118883 KP118884 KP118885 KP118887 KP118888 KP118891 KP118893 MK032177 MK032179 MK032180 ACD93207.1 ACT55271.1 AGY56144.1 AJP16725.1 AJP16734.1 AJT47846.1 AJT47855.1 AJT47864.1 AJT47873.1 AJT47882.1 AJT47891.1 AJT47909.1 AJT47918.1 AJT47945.1 AJT47963.1 AZA07828.1 AZA07846.1 AZA07854.1 MK887092 QEY95697.1 MK887102 QEY95707.1 JX273191 AFS18111.1 JX273197 AFS18117.1 KX258195 ANY58998.1 KF834569 KM365468 AIB54160.1 AJP08841.1 FJ849797 FJ849800 ACZ67619.1 ACZ67622.1 KU361187 ANO81447.1 M21515 KX272465 APY20698.1 KX219801 APV46188.1 FJ965349 ACR55678.1 AY646283 AAT70776.1 AY702975 KJ425496 KP343691 KT852992 KR608272 KU356856 KT002187 AAU14252.1 AHX25996.1 AKJ66257.1 AMD40267.1 AMH39309.1 ANI21153.1 AOG62366.1 EF602451 KX252791 ABQ84857.1 APY26731.1 GQ174477 ACS45209.1 GQ174478 ACS45210.1 KT736032 AMK51952.1 AF470628 AAO33466.1 KJ940499 KR265090 AJP08751.1 ART35337.1 MK581200 MK581201 QDQ69099.1 QDQ69112.1 KX252773 APY23492.1 GQ174483 ACS45215.1 KF574761 KF558356 KX236008 KX275391 KX372250 AGZ20166.1 AHJ77582.1 APY26811.1 APZ73820.1 AQY03972.1 GQ227603 ACT55272.1 FJ849820 ACZ67642.1 KP118886 AJT47900.1 FJ919617 ACQ91091.1 FJ904721 ADA83551.1 AF202999 AF206266 AF206267 AF206268 AF206269 AF206270 AF206271 JX273195 AFS18115.1 JX014375 AFN88521.1 KY626045 ARJ35795.1 AY325728 AY325729 GQ174486 FJ904720 FJ904723 GQ504724 GQ504725 JQ693036 KJ425504 KJ425508 KY805845 KY273667 MH539771 MH539772 MK728875 MK937830 MK937833 AAP86204.1 ACS45218.1 ADA83541.1 ADA83571.1 ADP06501.1 ADP06511.1 AFZ88831.1 AHX26068.1 AHX26104.1 ARS23143.1 ASF20084.1 AZP23934.1 AZP23944.1 QCE31537.1 QDA76313.1 QDA76340.1 AF363609 AF343343 AF286184 DQ834384 AY851295 AY044184 AF286185 KT946798 MG517474 AMR60484.1 AYD78153.1 MK887112 QEY95717.1 DQ490210 DQ490211 DQ490213 DQ490218 JN983807 KF460437 KF931628 ABF61506.1 ABF61514.1 ABF61527.1 ABF61567.1 AFK30957.1 AIG54287.1 AIG54299.1 AY514485 AAS00084.1 AY942743 AAX39762.1 DQ490205 ABF61469.1 EU817497 ACJ12838.1 MH151329 QAV53737.1 AY325735 KC013541 KC119407 AAP86210.1 AFY11702.1 AFY11711.1 JF705860 JF699749 AEO86772.1 AEO88932.1 FJ849811 ACZ67633.1 KX219796 APV46143.1 GQ427173 ACV87238.1 EU780078 ACE88707.2 EU714029 ACH72808.1 KF411041 AGZ89685.1 AY302747 AAP68695.1 KP036502 AJP16716.1 AF363599 AAK50026.1 MK071267 QCS40644.1 AY302742 AY302743 AY325732 AY846835 AY856347 EF602437 EF602438 EF602439 EF602443 EU714028 EU822341 FJ807652 FJ888351 FJ919614 FJ849808 GU393335 JF330899 JF828981 JX273198 KF188434 KJ425486 KJ425487 KJ425488 KJ425490 KJ425491 KJ425492 KJ425493 KJ425494 KJ425495 KJ425497 KJ425498 KJ425499 KJ425500 KJ425501 KJ425502 KJ425503 KJ425505 KJ425506 KJ425507 KJ425509 KJ425510 KJ425511 KJ425512 KJ435284 KT203557 MG763935 MK937829 MK887093 MK887097 MK887099 MK887106 AAP68690.1 AAP68691.1 AAP86207.1 AAW56626.1 AAW83033.1 ABQ84746.1 ABQ84754.1 ABQ84762.1 ABQ84794.1 ACH72797.1 ACJ50203.1 ACO37570.1 ACQ55234.1 ACQ91088.1 ACZ67630.1 ADV71781.1 ADY62556.1 AEP84750.1 AFS18118.1 AGW24531.1 AHX25906.1 AHX25915.1 AHX25924.1 AHX25942.1 AHX25951.1 AHX25960.1 AHX25969.1 AHX25978.1 AHX25987.1 AHX26005.1 AHX26014.1 AHX26023.1 AHX26032.1 AHX26041.1 AHX26050.1 AHX26059.1 AHX26077.1 AHX26086.1 AHX26095.1 AHX26113.1 AHX26122.1 AHX26131.1 AHX26140.1 AHX26158.1 ALE71335.1 AVX27616.1 QDA76304.1 QEY95698.1 QEY95702.1 QEY95704.1 QEY95711.1 AY028295 AF203004 AF322367 JX014367 KY407556 KY407558 AFN88513.1 AQY61309.1 AQY61327.1 MG913342 AXQ05194.1 FJ849796 FJ849805 FJ849807 FJ849826 JQ801377 ACZ67618.1 ACZ67627.1 ACZ67629.1 ACZ67648.1 AFJ24941.1 DQ490221 ABF61583.1 KX302867 APZ73719.1 KF975407 KJ669722 KJ940498 KR265091 AIB54164.1 AJP08745.1 AJP08750.1 ART35345.1 KF975408 KR265087 AIB54165.1 ART35313.1 AF363601 AF363604 AY325727 EF602457 GQ504723 KX236001 MK937828 MK887103 MK887109 AAK50028.1 AAK50031.1 AAP86202.1 ABQ84905.1 ADP06488.1 APY26749.1 QDA76295.1 QEY95708.1 QEY95714.1 MG913343 AXQ05205.1
AHB63483.1 AHB63510.1 FJ976459 ACR16060.1 EU826085 ACE95721.1 MH151328 QAV53736.1 FJ976465 ACR16066.1 FJ976467 ACR16068.1 FJ976460 ACR16061.1 FJ976461 ACR16062.1 MK887107 QEY95712.1 FJ849823 ACZ67645.1 GU393332 ADV71743.1 JX565012 MH924835 MK142676 AFV92602.1 AZA07518.1 QCX41899.1 FJ965348 GQ174475 ACR55677.1 ACS45207.1 GQ174481 ACS45213.1 EF602444 ABQ84802.1 MK887111 QEY95716.1 KY626044 ARJ35785.1 JX565011 AFV92601.1 FJ807653 GQ179645 JF893452 KF663560 ACO37578.1 ACS45205.1 AEJ80242.1 AHB74003.1 GQ174474 HQ283475 HQ850618 ACS45206.1 ADT91599.1 AEB00719.1 KT886454 AMO03296.1 KM213963 AIY51832.1 MK887104 MK887108 QEY95709.1 QEY95713.1 HQ141058 ADW77296.1 DQ490214 ABF61535.1 EF602450 KY799582 ABQ84850.1 AVI69489.1 GQ174485 ACS45217.1 EU714034 GQ227602 KF377577 KP036503 KP036504 KP118880 KP118881 KP118882 KP118883 KP118884 KP118885 KP118887 KP118888 KP118891 KP118893 MK032177 MK032179 MK032180 ACD93207.1 ACT55271.1 AGY56144.1 AJP16725.1 AJP16734.1 AJT47846.1 AJT47855.1 AJT47864.1 AJT47873.1 AJT47882.1 AJT47891.1 AJT47909.1 AJT47918.1 AJT47945.1 AJT47963.1 AZA07828.1 AZA07846.1 AZA07854.1 MK887092 QEY95697.1 MK887102 QEY95707.1 JX273191 AFS18111.1 JX273197 AFS18117.1 KX258195 ANY58998.1 KF834569 KM365468 AIB54160.1 AJP08841.1 FJ849797 FJ849800 ACZ67619.1 ACZ67622.1 KU361187 ANO81447.1 M21515 KX272465 APY20698.1 KX219801 APV46188.1 FJ965349 ACR55678.1 AY646283 AAT70776.1 AY702975 KJ425496 KP343691 KT852992 KR608272 KU356856 KT002187 AAU14252.1 AHX25996.1 AKJ66257.1 AMD40267.1 AMH39309.1 ANI21153.1 AOG62366.1 EF602451 KX252791 ABQ84857.1 APY26731.1 GQ174477 ACS45209.1 GQ174478 ACS45210.1 KT736032 AMK51952.1 AF470628 AAO33466.1 KJ940499 KR265090 AJP08751.1 ART35337.1 MK581200 MK581201 QDQ69099.1 QDQ69112.1 KX252773 APY23492.1 GQ174483 ACS45215.1 KF574761 KF558356 KX236008 KX275391 KX372250 AGZ20166.1 AHJ77582.1 APY26811.1 APZ73820.1 AQY03972.1 GQ227603 ACT55272.1 FJ849820 ACZ67642.1 KP118886 AJT47900.1 FJ919617 ACQ91091.1 FJ904721 ADA83551.1 AF202999 AF206266 AF206267 AF206268 AF206269 AF206270 AF206271 JX273195 AFS18115.1 JX014375 AFN88521.1 KY626045 ARJ35795.1 AY325728 AY325729 GQ174486 FJ904720 FJ904723 GQ504724 GQ504725 JQ693036 KJ425504 KJ425508 KY805845 KY273667 MH539771 MH539772 MK728875 MK937830 MK937833 AAP86204.1 ACS45218.1 ADA83541.1 ADA83571.1 ADP06501.1 ADP06511.1 AFZ88831.1 AHX26068.1 AHX26104.1 ARS23143.1 ASF20084.1 AZP23934.1 AZP23944.1 QCE31537.1 QDA76313.1 QDA76340.1 AF363609 AF343343 AF286184 DQ834384 AY851295 AY044184 AF286185 KT946798 MG517474 AMR60484.1 AYD78153.1 MK887112 QEY95717.1 DQ490210 DQ490211 DQ490213 DQ490218 JN983807 KF460437 KF931628 ABF61506.1 ABF61514.1 ABF61527.1 ABF61567.1 AFK30957.1 AIG54287.1 AIG54299.1 AY514485 AAS00084.1 AY942743 AAX39762.1 DQ490205 ABF61469.1 EU817497 ACJ12838.1 MH151329 QAV53737.1 AY325735 KC013541 KC119407 AAP86210.1 AFY11702.1 AFY11711.1 JF705860 JF699749 AEO86772.1 AEO88932.1 FJ849811 ACZ67633.1 KX219796 APV46143.1 GQ427173 ACV87238.1 EU780078 ACE88707.2 EU714029 ACH72808.1 KF411041 AGZ89685.1 AY302747 AAP68695.1 KP036502 AJP16716.1 AF363599 AAK50026.1 MK071267 QCS40644.1 AY302742 AY302743 AY325732 AY846835 AY856347 EF602437 EF602438 EF602439 EF602443 EU714028 EU822341 FJ807652 FJ888351 FJ919614 FJ849808 GU393335 JF330899 JF828981 JX273198 KF188434 KJ425486 KJ425487 KJ425488 KJ425490 KJ425491 KJ425492 KJ425493 KJ425494 KJ425495 KJ425497 KJ425498 KJ425499 KJ425500 KJ425501 KJ425502 KJ425503 KJ425505 KJ425506 KJ425507 KJ425509 KJ425510 KJ425511 KJ425512 KJ435284 KT203557 MG763935 MK937829 MK887093 MK887097 MK887099 MK887106 AAP68690.1 AAP68691.1 AAP86207.1 AAW56626.1 AAW83033.1 ABQ84746.1 ABQ84754.1 ABQ84762.1 ABQ84794.1 ACH72797.1 ACJ50203.1 ACO37570.1 ACQ55234.1 ACQ91088.1 ACZ67630.1 ADV71781.1 ADY62556.1 AEP84750.1 AFS18118.1 AGW24531.1 AHX25906.1 AHX25915.1 AHX25924.1 AHX25942.1 AHX25951.1 AHX25960.1 AHX25969.1 AHX25978.1 AHX25987.1 AHX26005.1 AHX26014.1 AHX26023.1 AHX26032.1 AHX26041.1 AHX26050.1 AHX26059.1 AHX26077.1 AHX26086.1 AHX26095.1 AHX26113.1 AHX26122.1 AHX26131.1 AHX26140.1 AHX26158.1 ALE71335.1 AVX27616.1 QDA76304.1 QEY95698.1 QEY95702.1 QEY95704.1 QEY95711.1 AY028295 AF203004 AF322367 JX014367 KY407556 KY407558 AFN88513.1 AQY61309.1 AQY61327.1 MG913342 AXQ05194.1 FJ849796 FJ849805 FJ849807 FJ849826 JQ801377 ACZ67618.1 ACZ67627.1 ACZ67629.1 ACZ67648.1 AFJ24941.1 DQ490221 ABF61583.1 KX302867 APZ73719.1 KF975407 KJ669722 KJ940498 KR265091 AIB54164.1 AJP08745.1 AJP08750.1 ART35345.1 KF975408 KR265087 AIB54165.1 ART35313.1 AF363601 AF363604 AY325727 EF602457 GQ504723 KX236001 MK937828 MK887103 MK887109 AAK50028.1 AAK50031.1 AAP86202.1 ABQ84905.1 ADP06488.1 APY26749.1 QDA76295.1 QEY95708.1 QEY95714.1 MG913343 AXQ05205.1
Proteomes
UP000125413
UP000134696
UP000101112
UP000126730
UP000160499
UP000156427
+ More
UP000170434 UP000107262 UP000133437 UP000174764 UP000100522 UP000105510 UP000108163 UP000109170 UP000111271 UP000121165 UP000124537 UP000136129 UP000142905 UP000146859 UP000147307 UP000149934 UP000170218 UP000118234 UP000102195 UP000138732 UP000139325 UP000159001 UP000107304 UP000102415 UP000166017 UP000169726 UP000098752 UP000107291 UP000114242 UP000117985 UP000155946 UP000165485 UP000007642 UP000096468 UP000099500 UP000123393 UP000128029 UP000160278 UP000107830 UP000119449 UP000165290 UP000172963 UP000123786 UP000139954 UP000096908 UP000105210 UP000097868 UP000102121 UP000102163 UP000103149 UP000106511 UP000108224 UP000113245 UP000113896 UP000113939 UP000118008 UP000120286 UP000120642 UP000132365 UP000136241 UP000136440 UP000138146 UP000146865 UP000149404 UP000155572 UP000158124 UP000158949 UP000159206 UP000163887 UP000165788 UP000166253 UP000167410 UP000170189 UP000170844 UP000173109 UP000173215 UP000180343 UP000132955
UP000170434 UP000107262 UP000133437 UP000174764 UP000100522 UP000105510 UP000108163 UP000109170 UP000111271 UP000121165 UP000124537 UP000136129 UP000142905 UP000146859 UP000147307 UP000149934 UP000170218 UP000118234 UP000102195 UP000138732 UP000139325 UP000159001 UP000107304 UP000102415 UP000166017 UP000169726 UP000098752 UP000107291 UP000114242 UP000117985 UP000155946 UP000165485 UP000007642 UP000096468 UP000099500 UP000123393 UP000128029 UP000160278 UP000107830 UP000119449 UP000165290 UP000172963 UP000123786 UP000139954 UP000096908 UP000105210 UP000097868 UP000102121 UP000102163 UP000103149 UP000106511 UP000108224 UP000113245 UP000113896 UP000113939 UP000118008 UP000120286 UP000120642 UP000132365 UP000136241 UP000136440 UP000138146 UP000146865 UP000149404 UP000155572 UP000158124 UP000158949 UP000159206 UP000163887 UP000165788 UP000166253 UP000167410 UP000170189 UP000170844 UP000173109 UP000173215 UP000180343 UP000132955
Pfam
PF01635
Corona_M
ProteinModelPortal
B2BW35
V5TGF3
V5TFF2
C5IZI4
B3VZS5
A0A481NU89
+ More
C5IZJ0 C5IZJ2 C5IZI5 C5IZI6 A0A5J6RCQ6 G4RJ98 G3C785 K7RWV4 C5IUA3 C6KCX8 A5Z0Z4 A0A5J6RC67 A0A1W6AWK4 K7S5A6 C1KAC3 C6KCX1 A0A140HEW9 A0A0A1G0F9 A0A5J6RDV5 E9LUF4 A4ZCQ8 A5Z142 C6KCY2 B3GPG8 A0A5J6RDK2 A0A5J6RDL5 J9WMJ2 J9WRY4 A0A1B2CWH4 A0A0U1WJZ5 G4RJ75 A0A1B0Z544 P12649 A0A1P8SFM3 A0A1P8FCL8 C5IUA4 Q6DTU8 Q64FZ0 A5Z149 C6KCX4 C6KCX5 A0A140DP47 Q80RY9 A0A1L1XGR3 A0A516UWB7 A0A1P8SMJ8 C6KCY0 U5U9J5 C7DY52 G4RJ95 A0A0D4CG32 C4PE61 F4MIY0 Q9J3N8 J9WLU4 I7B3F0 A0A1W6AWL0 Q6VUR2 P69606 P69607 A0A142IJS1 A0A5J6RDM6 A4ZCM9 Q6R4P1 Q58J43 A4ZCJ2 C7S863 A0A481NUK2 Q7T9P6 G3FEJ2 G4RJ86 A0A1P8FCE8 C9DST0 B3VN11 C7S7Z4 U5YDU7 Q7TEH1 A0A0C5GXD2 Q91SA2 A0A4V6MBA0 Q6W9D6 P69604 P69603 P69605 I7AZ79 A0A346M350 G4RJA1 A4ZCV6 A0A1P8VF79 A0A0U1UZF0 A0A0U1WK58 Q910H8 A0A346M361
C5IZJ0 C5IZJ2 C5IZI5 C5IZI6 A0A5J6RCQ6 G4RJ98 G3C785 K7RWV4 C5IUA3 C6KCX8 A5Z0Z4 A0A5J6RC67 A0A1W6AWK4 K7S5A6 C1KAC3 C6KCX1 A0A140HEW9 A0A0A1G0F9 A0A5J6RDV5 E9LUF4 A4ZCQ8 A5Z142 C6KCY2 B3GPG8 A0A5J6RDK2 A0A5J6RDL5 J9WMJ2 J9WRY4 A0A1B2CWH4 A0A0U1WJZ5 G4RJ75 A0A1B0Z544 P12649 A0A1P8SFM3 A0A1P8FCL8 C5IUA4 Q6DTU8 Q64FZ0 A5Z149 C6KCX4 C6KCX5 A0A140DP47 Q80RY9 A0A1L1XGR3 A0A516UWB7 A0A1P8SMJ8 C6KCY0 U5U9J5 C7DY52 G4RJ95 A0A0D4CG32 C4PE61 F4MIY0 Q9J3N8 J9WLU4 I7B3F0 A0A1W6AWL0 Q6VUR2 P69606 P69607 A0A142IJS1 A0A5J6RDM6 A4ZCM9 Q6R4P1 Q58J43 A4ZCJ2 C7S863 A0A481NUK2 Q7T9P6 G3FEJ2 G4RJ86 A0A1P8FCE8 C9DST0 B3VN11 C7S7Z4 U5YDU7 Q7TEH1 A0A0C5GXD2 Q91SA2 A0A4V6MBA0 Q6W9D6 P69604 P69603 P69605 I7AZ79 A0A346M350 G4RJA1 A4ZCV6 A0A1P8VF79 A0A0U1UZF0 A0A0U1WK58 Q910H8 A0A346M361
PDB
5H3O
E-value=1.26658,
Score=71
Ontologies
KEGG
GO
Subcellular Location
From MSLVP
Host nucleus. Host cytoplasm.
From Uniprot
Host Golgi apparatus membrane
Virion membrane
Virion membrane
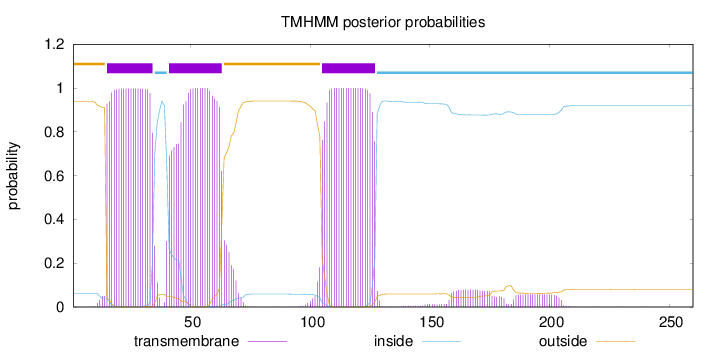
Topology
Length:
260
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
68.2689800000001
Exp number, first 60 AAs:
38.67324
Total prob of N-in:
0.06185
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 34
inside
35 - 40
TMhelix
41 - 63
outside
64 - 104
TMhelix
105 - 127
inside
128 - 260
Population Genetic Test Statistics
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 |
Protein |
YP_001876439.1 |
|
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.