Strain
CANADA_AVIAN_2007_NC_010800
(Region: Canada; Strain: Turkey coronavirus, complete genome.; Date: 2007)
Gene
spike protein
Description
Annotated in NCBI,
spike protein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGTTGGTGCAGTTGTGTTGCCTGACGATAATCTTGTCACTTGTATGTAATGTGAGTTCTATTCCCACAATTTGTCTCAATGCTACTGTAGGTAGTGGTGTGTGTCTACCTTGCACAAACATACCAGAGTATGCAGATGGTAAAAATGGGGGTGAAGCCCTAGATTTTTATAGTCCAGATGTTATGAGACCGCCAGATGGCGCATATATACAATCCGGTTATTATGAACCACTTTTCACAGGTTGCTTTAATCAAACTAATCAAACTGATAGCACCTGTAGAAATGGGCTTTATAAAGGGTCGCCAGGTAATTTTACTATACAAGGTGATTTTCTTAAAAATTATGATGCTGTTGGCATAATGTTTTGGTGGGGTTTAGCTACTAATGTGGGTAAAGTAACACCACCCAATGATCCTACTTTTAATTTAACATGGGGCAACTTTTTTCTTAATTCTAAGAATTTTACAGGTTTCCATAAAGTTAAAAGTGTCATATTTATTGCCACTGGAGATATCTTCGTAAATGGCGTTTTAATGGGTTTTTATAATCTAAATTTTACACAAACCTTAACAATTTGGTTAGCACAGTGTGTTGGCACAATGAAGGTTGTTATTTTACGTAATAGTAATGCTCTAGTTCATTTTTCGGCTGGCAACGTAGTTGCTTTCGAACCCTGCACAGGAGACACTACTATTAATAAGTTACGTTGCGCTTATCAGCAATTTAATTTTTCTACAGGATTTTATGACATAGATACTTTTGTACCTGTGACATCTAATATTACATATTTACCTTACCCAGATTTAAAAGACAATACTGGTCAAGAGGTATGTGACTTTTATGTAGCTCTTAAAGGAGATCATGTTATTTACAACCAAAGTTGTGTAGACTCTAAGTACCCATTCTTTAAATTAAAGTGTAATAATACTTATTCATGGGATAATGATAAGTTGTGTATTTTAGGGTCTGAAATATATGTGCCGGGGTATAATGTTTACACTAATACGCAACATTCATATGCTGGCACTATACCGAATTATCCAACTTGTTCTTCCAAAGGTCTTAGTTTGGAAAATATTTATAATAATATAGGTTTCAATAGATTTTGTGTAACTAAATTGTTGACGACTCGTGATATATCATCTATAACCCAATACACTTGTGTTTATGTCGTTAGTGTAGAAGGTTTTGATAGTTATAATCAAGGTATTGTATACAGTTATGTTGGTGTAAACTTTGATTTTGGTACAGCTATGTATAGTGTGAAAACTGCACCTATGCAGTTCATCTATACAGAGCAATGTCATAAGTATAATATTTACAACATAAGAGGTGTTGGTCAAATATTAAATGTCACTGGTAAAGATAATACTACACTTGTAGACGGTGGTCTAGTAATAACTAGTGGCAGTGGCTTATTGGCTTTTCGTAATAATGGAAGTCTTTTTAGTGTACAACCTTGCCGGACAGTTTCAACTCAAGCTGTAATTGTTAATGGGAGTTTTGTAGGCTTGTTTGTTCCTGCATCTTGTTGGTTAGCCGATCAATTTAATCTAGGTAACCACACAGAATATGTAGATGGTGGTTGTCTTATTAGTAATAAGACTATTAGTAGAAAGCGTAGGTCTACTAACGCTGTTTATACTGGTGAATGCACAGGTTTATTTGCAGCTATGGGTACATCTTGTATTTATAGTAATGGCACAGTTCTTAATAGAACATTACCACAACCATCTGTTCAAACTAGTGTGCAACCATTACTAGGTGTTGTAGCAAATGTGTCTATTCCAAAAGAATTGACACTTGCTGTAACCACAGAATACTTACAAACTCGTTATCAGAAAGTTACTATTGATTGTGCTAAATATGTCTGTGGTGATTCTACAAGGTGCCGCACGTTGCTGCAGCAGTATGGTTCTTTTTGCCAGAGTGTTAATGCTATATTAAGTGGTGTTAATGAGAATGAAGATAATGGTTTACTGCAATATGCGGAAGCTATTAATACAGGTTACACACTTAATTTTACTGGTTATAATGTATCCAATTTGGGTGGGTTTGATTTGTCCCTTGTTCTACCCAAAAATCTTACTTTGGCACAAAATCAAGGTAGGTCTACTATTGAAGACTTACTTTTTGACAAAGTTGTTACTCTAGGTGTTAGTGAAGTTGATCAGAATTATGATAAATGTATAGCATCTAGAGGTGGTTCTTTTACTAATTTAGCAGATTTAACTTGTGCTCAGTTTTACAATGGTGTCATGGTGCTCCCTGGTGTTGTTGACCCTGGTCTAATGGCAGTGTACACCGGCTCTTTAATAGGTGGTATGGCGCTCGGAGGCATTACAGCAGCGGCATCCATACCATTTGCTACTCAAATACAAGCACGTGTCAATTATTTAGCGTTGACACAATCAGTGCTTTTAGATAACCAAAATTTGATAGCAAATTCCTTTAATAAGGCCCTTAAAGGTATACAGTCAGCATTAGACACTGTTTCCCAGGGGTTTGTTGAAGTAGCTAGAGGTTTTGAGAGCGTAACTGTAGCACTTAATAAAGTTCAGGATGTGGTTAACACACATTCCGACATATTAAATAAACTTATGGCTCAGCTTAGTGTTAATTTTGGGGCTGTTTCATCTTCTCTTAATGAGATATACCTAAAATTAGACCAGATTAATGCAGATGCTCAGGTTGATAGGTTAATTACTGGTAGGTTGACTGCATTATCAACTTACGTAGCATCTCTTCAATTAGCAGCTTATAAGGCTGACCAGTCAAGACGCTTGGCTCTACAGAAAGTAGAGGAGTGTGTTAAGTCGCAATCCATGCGATATGGTTTCTGTGGTAATGGTAGCCATGTTCTAACAATACCACAGAGTGCTCCTAATGGAATATTCTTTATCCACTACACGTACCAACCCACTTCTTATGTTACAATTGAAGCAGTGCCTGGTCTATGTGTAACAACACCTTCTGGAAAGTATGGTGTAATGCCCAAGACTGGAAGTGGCATCATATTTAGACAGAATGATACTTTCTTTGTCACTTCCACACAGTTATATGAGCCTAAGCTTTTATCATATTCAGAAGTGGTCAATTTAACATCTTGTGAAGCTAATTATTACAATGTTAGTGAAAGTGAAACACCCTTTCAACCAGCATTGCCTAATTTCGATGACGAGTTTAATGACATTTTTACTGAGTTAAATACCTCAAAAGATGCCATTAAAAATATCAGTTCCAATTTTAACTACACAATTCCCATCCTAAATCTTGAGGATGAGATTTGTAGATTGAATCACTCCATTAACACACTATATAATTATAGTGATGTTATTGATAAAATTAACCAGGGTCTTAATGACACCTTTATTAATCTTGAACAACTTAATAAAATTACACGTTACATTAAGTGGCCTTGGTATGTTTGGTTAGCAATTGGTTTTGCTTGTTTAATATTCATCTTAATATTAGGATGGGTGTTTTTCATGACTGGTTGTTGTGGTTGTTGTTGCGGATGCTTTGGCATTATTCCGCTAATGAGTAAGTGTGGTAAGAAATCCTCTTACTACACGACTTTTGATAATGATGTGGTGTATAACAAAACAGACCTAAAAAGTCTGTTTAATGATTCAAACTCCAACATCTTTTCTAATAGTGTTAATTCTTCTTTGGTTTAA
Protein
MLVQLCCLTIILSLVCNVSSIPTICLNATVGSGVCLPCTNIPEYADGKNGGEALDFYSPDVMRPPDGAYIQSGYYEPLFTGCFNQTNQTDSTCRNGLYKGSPGNFTIQGDFLKNYDAVGIMFWWGLATNVGKVTPPNDPTFNLTWGNFFLNSKNFTGFHKVKSVIFIATGDIFVNGVLMGFYNLNFTQTLTIWLAQCVGTMKVVILRNSNALVHFSAGNVVAFEPCTGDTTINKLRCAYQQFNFSTGFYDIDTFVPVTSNITYLPYPDLKDNTGQEVCDFYVALKGDHVIYNQSCVDSKYPFFKLKCNNTYSWDNDKLCILGSEIYVPGYNVYTNTQHSYAGTIPNYPTCSSKGLSLENIYNNIGFNRFCVTKLLTTRDISSITQYTCVYVVSVEGFDSYNQGIVYSYVGVNFDFGTAMYSVKTAPMQFIYTEQCHKYNIYNIRGVGQILNVTGKDNTTLVDGGLVITSGSGLLAFRNNGSLFSVQPCRTVSTQAVIVNGSFVGLFVPASCWLADQFNLGNHTEYVDGGCLISNKTISRKRRSTNAVYTGECTGLFAAMGTSCIYSNGTVLNRTLPQPSVQTSVQPLLGVVANVSIPKELTLAVTTEYLQTRYQKVTIDCAKYVCGDSTRCRTLLQQYGSFCQSVNAILSGVNENEDNGLLQYAEAINTGYTLNFTGYNVSNLGGFDLSLVLPKNLTLAQNQGRSTIEDLLFDKVVTLGVSEVDQNYDKCIASRGGSFTNLADLTCAQFYNGVMVLPGVVDPGLMAVYTGSLIGGMALGGITAAASIPFATQIQARVNYLALTQSVLLDNQNLIANSFNKALKGIQSALDTVSQGFVEVARGFESVTVALNKVQDVVNTHSDILNKLMAQLSVNFGAVSSSLNEIYLKLDQINADAQVDRLITGRLTALSTYVASLQLAAYKADQSRRLALQKVEECVKSQSMRYGFCGNGSHVLTIPQSAPNGIFFIHYTYQPTSYVTIEAVPGLCVTTPSGKYGVMPKTGSGIIFRQNDTFFVTSTQLYEPKLLSYSEVVNLTSCEANYYNVSESETPFQPALPNFDDEFNDIFTELNTSKDAIKNISSNFNYTIPILNLEDEICRLNHSINTLYNYSDVIDKINQGLNDTFINLEQLNKITRYIKWPWYVWLAIGFACLIFILILGWVFFMTGCCGCCCGCFGIIPLMSKCGKKSSYYTTFDNDVVYNKTDLKSLFNDSNSNIFSNSVNSSLV
Summary
Function
Spike protein S1: attaches the virion to the host cell membrane by interacting with sialic acids, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide after S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
attaches the virion to the host cell membrane by interacting with sialic acids, initiating the infection.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide after S2 cleavage occurring upon virus endocytosis.
Spike protein S2': Acts as a viral fusion peptide after S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
attaches the virion to the host cell membrane by interacting with sialic acids, initiating the infection.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide after S2 cleavage occurring upon virus endocytosis.
Subunit
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes.
Similarity
Belongs to the gammacoronaviruses spike protein family.
Keywords
Cleavage on pair of basic residues
Coiled coil
Fusion of virus membrane with host endosomal membrane
Fusion of virus membrane with host membrane
Glycoprotein
Host membrane
Host-virus interaction
Membrane
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Viral penetration into host cytoplasm
Virion
Virulence
Virus endocytosis by host
Virus entry into host cell
Feature
chain Spike protein S2
Uniprot
B3FHU5
C9DSU8
A0A0N6YPJ4
D0ECU0
A0A0N6YR14
D3X6W4
+ More
D3X6W5 D3X6W3 C9DSV9 A0A0N6YR01 A0A0N6YPZ1 A0A0N6YPJ7 A0A0N6YPI7 A0A0N6YPK1 A0A0N6YPH6 A0A0N6YSM3 A0A0N6YPJ5 D0ECU1 A0A0N6YSN1 B3TD48 A0A0N6YPX8 A0A0N6YPY3 A0A0N6YPK6 D3X6W6 A0A0N6YR06 D0ECU2 A0A0N6YQK0 A0A0N6YPI3 C9DST7 A0A0N6W221 A0A0N6YQK7 A0A0N6YPD0 A0A0N6YSN7 B3TD61 A0A0N6W222 A0A0N6YPD8 Q7T5V2 Q7T5V3 A0A3S7SW51 U4DCU7 A0A4Y5QS01 A0A0F6YS09 A0A0S2ZWY7 E6YDV7 D9YMZ5 E6YDV6 A0A2Z5D876 P12722 P05135
D3X6W5 D3X6W3 C9DSV9 A0A0N6YR01 A0A0N6YPZ1 A0A0N6YPJ7 A0A0N6YPI7 A0A0N6YPK1 A0A0N6YPH6 A0A0N6YSM3 A0A0N6YPJ5 D0ECU1 A0A0N6YSN1 B3TD48 A0A0N6YPX8 A0A0N6YPY3 A0A0N6YPK6 D3X6W6 A0A0N6YR06 D0ECU2 A0A0N6YQK0 A0A0N6YPI3 C9DST7 A0A0N6W221 A0A0N6YQK7 A0A0N6YPD0 A0A0N6YSN7 B3TD61 A0A0N6W222 A0A0N6YPD8 Q7T5V2 Q7T5V3 A0A3S7SW51 U4DCU7 A0A4Y5QS01 A0A0F6YS09 A0A0S2ZWY7 E6YDV7 D9YMZ5 E6YDV6 A0A2Z5D876 P12722 P05135
EMBL
EU095850
GQ427173
ABW81427.1
ACV87236.1
GQ427175
ACV87265.1
+ More
KF652240 AIL92614.1 GQ469644 ACX30000.1 KF652235 AIL92609.1 GU213200 ADD14081.1 GU213201 ADD14082.1 GU213199 ADD14080.1 GQ427176 GQ469643 ACV87276.1 ACX29999.1 KF652218 AIL92592.1 KF652238 AIL92612.1 KF652223 AIL92597.1 KF652221 AIL92595.1 KF652239 AIL92613.1 KF652222 AIL92596.1 KF652219 AIL92593.1 KF652230 AIL92604.1 GQ469645 ACX30001.1 KF652228 AIL92602.1 EU022525 ABW75125.1 KF652220 AIL92594.1 KF652229 AIL92603.1 KF652232 AIL92606.1 GU213202 ADD14083.1 KF652227 AIL92601.1 GQ469646 ACX30002.1 KF652225 AIL92599.1 KF652231 AIL92605.1 GQ427174 ACV87254.1 KF652226 AIL92600.1 KF652234 AIL92608.1 KF652224 AIL92598.1 KF652237 AIL92611.1 EU022526 ABW75138.1 KF652236 AIL92610.1 KF652233 AIL92607.1 AY342357 AAQ20922.1 AY342356 AAQ20921.1 MG765535 AYC44319.1 HF544506 LN610099 CCN27367.1 CEK09108.1 MK142676 QCX41896.1 KM454473 AKF17724.1 KR822424 ALQ43515.1 FN545819 CBB12677.1 GQ411201 ADK88938.1 FN434203 CBA13338.1 MH021175 AXB38894.1 X15832 X04723
KF652240 AIL92614.1 GQ469644 ACX30000.1 KF652235 AIL92609.1 GU213200 ADD14081.1 GU213201 ADD14082.1 GU213199 ADD14080.1 GQ427176 GQ469643 ACV87276.1 ACX29999.1 KF652218 AIL92592.1 KF652238 AIL92612.1 KF652223 AIL92597.1 KF652221 AIL92595.1 KF652239 AIL92613.1 KF652222 AIL92596.1 KF652219 AIL92593.1 KF652230 AIL92604.1 GQ469645 ACX30001.1 KF652228 AIL92602.1 EU022525 ABW75125.1 KF652220 AIL92594.1 KF652229 AIL92603.1 KF652232 AIL92606.1 GU213202 ADD14083.1 KF652227 AIL92601.1 GQ469646 ACX30002.1 KF652225 AIL92599.1 KF652231 AIL92605.1 GQ427174 ACV87254.1 KF652226 AIL92600.1 KF652234 AIL92608.1 KF652224 AIL92598.1 KF652237 AIL92611.1 EU022526 ABW75138.1 KF652236 AIL92610.1 KF652233 AIL92607.1 AY342357 AAQ20922.1 AY342356 AAQ20921.1 MG765535 AYC44319.1 HF544506 LN610099 CCN27367.1 CEK09108.1 MK142676 QCX41896.1 KM454473 AKF17724.1 KR822424 ALQ43515.1 FN545819 CBB12677.1 GQ411201 ADK88938.1 FN434203 CBA13338.1 MH021175 AXB38894.1 X15832 X04723
Proteomes
Gene 3D
ProteinModelPortal
B3FHU5
C9DSU8
A0A0N6YPJ4
D0ECU0
A0A0N6YR14
D3X6W4
+ More
D3X6W5 D3X6W3 C9DSV9 A0A0N6YR01 A0A0N6YPZ1 A0A0N6YPJ7 A0A0N6YPI7 A0A0N6YPK1 A0A0N6YPH6 A0A0N6YSM3 A0A0N6YPJ5 D0ECU1 A0A0N6YSN1 B3TD48 A0A0N6YPX8 A0A0N6YPY3 A0A0N6YPK6 D3X6W6 A0A0N6YR06 D0ECU2 A0A0N6YQK0 A0A0N6YPI3 C9DST7 A0A0N6W221 A0A0N6YQK7 A0A0N6YPD0 A0A0N6YSN7 B3TD61 A0A0N6W222 A0A0N6YPD8 Q7T5V2 Q7T5V3 A0A3S7SW51 U4DCU7 A0A4Y5QS01 A0A0F6YS09 A0A0S2ZWY7 E6YDV7 D9YMZ5 E6YDV6 A0A2Z5D876 P12722 P05135
D3X6W5 D3X6W3 C9DSV9 A0A0N6YR01 A0A0N6YPZ1 A0A0N6YPJ7 A0A0N6YPI7 A0A0N6YPK1 A0A0N6YPH6 A0A0N6YSM3 A0A0N6YPJ5 D0ECU1 A0A0N6YSN1 B3TD48 A0A0N6YPX8 A0A0N6YPY3 A0A0N6YPK6 D3X6W6 A0A0N6YR06 D0ECU2 A0A0N6YQK0 A0A0N6YPI3 C9DST7 A0A0N6W221 A0A0N6YQK7 A0A0N6YPD0 A0A0N6YSN7 B3TD61 A0A0N6W222 A0A0N6YPD8 Q7T5V2 Q7T5V3 A0A3S7SW51 U4DCU7 A0A4Y5QS01 A0A0F6YS09 A0A0S2ZWY7 E6YDV7 D9YMZ5 E6YDV6 A0A2Z5D876 P12722 P05135
PDB
6CV0
E-value=1.83774e-124,
Score=1146
Ontologies
KEGG
GO
GO:0061025 P:membrane fusion
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
Subcellular Location
From MSLVP
Capsid
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers may be transported to the plasma membrane, where they may mediate cell-cell fusion. S1 is not anchored to the viral envelope, but associates with the extravirion surface through its binding to S2. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers may be transported to the plasma membrane, where they may mediate cell-cell fusion. S1 is not anchored to the viral envelope, but associates with the extravirion surface through its binding to S2. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers may be transported to the plasma membrane, where they may mediate cell-cell fusion. S1 is not anchored to the viral envelope, but associates with the extravirion surface through its binding to S2. With evidence from 1 publications.
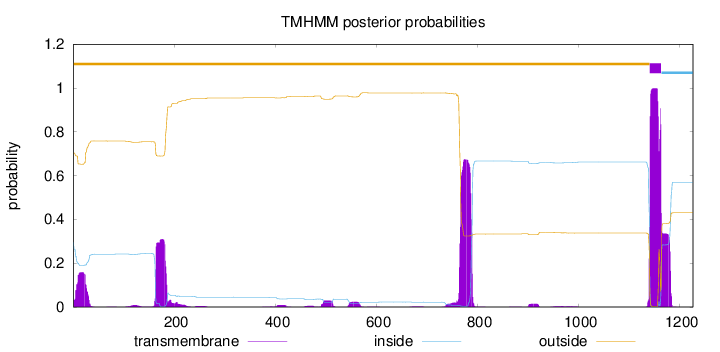
Topology
Length:
1226
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
57.42818
Exp number, first 60 AAs:
3.50987
Total prob of N-in:
0.29651
outside
1 - 1140
TMhelix
1141 - 1163
inside
1164 - 1226
Population Genetic Test Statistics
Pi
0.4156523
Theta
0.5114868
Tajima's D
-0.7705678
CLR
55.635335
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 |
Protein |
YP_001941166.1 |
|
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.