Strain
CHINA_AVIAN_2007_NC_016992
(Region: China: Hong Kong; Strain: Sparrow coronavirus HKU17, complete genome.; Date: 2007)
Gene
spike glycoprotein
Description
Annotated in NCBI,
spike glycoprotein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGCAGGGAGCTCTACTGATCACATTTATCGCAGTGGTGACCTCGTCGCCGCTAGCTGATAAAATACTTGATTTTCTAACTTTTCCTGAAGCACATGCCTATCTATTTCCGCAGTCACGCATGGTCAGGAATATAGAGGGCGGTGCTACTGAACGGTGTATGTACGTCCAAGAGGGCGGTTTCATACCAGACAACTTTACATTCCCTCAGTGGTTTGTACTCACCAACGACTCTACATTGCTTCAAGGAGAGTTAACAACGTCGCAGCCGCTAATTGTCAATGGGCACCTTTGCTCATGGCAATCTAACCAACAATATCGAAAATATTCCTTTAACACCTCTTGCCCACTTCCGGCTGACGGTTGTAACCCCACCTGGGGTAGTGGCAATCTTAAGTTGTTTAAGGGATGGAACAACACGAAAGGTCTTACTGACAATATCAGAATTAATGTCAACATATCCCAGTCGGATTATAGGACTTCTGCTGGGTCTGTTGGACTTGAATTAGAAGGAGGTGGTACTGTTAATATTACTTGTACTAACAGTTCCACGCCAGTCACAACATACTCACTACTACCTTGGGCTCGTTCCAGTGGTGAACCAATCTATTGCTTTGCCAATGTTTCATCACCTAGTCAAAGCTATATTGATTTTATGGGCATTCTCCCGCCTTTTGTGTCAGAGATTGCCTTCGATAGGAGTGGCAGTATCTATATTAATGGTTATAGATACTTCAAAATTTCGCCACTACTCAATGTAGAGCTTCAGCTCAACTTCAATCTAACCAGTGACCACTTCTCAGTCACTTGGTCTAATTACACTGAGGTACATTTAAACACTACCAATGGTTATATACATCAGATTAAATACTGCTATGACCCACTTGACAAACTGGCGTGTGAAATGAACACATTCCAATTGCCTGATGGAGTCTATCCATACACGCCCGCTCAACAGCAGGCTTTACCTGAAACATTCGTTACTACACCTGTGTATGCTAATCATACGACTGTTGTAGTACGCACTCAATACACAGTGAGTTCAACCGGAATTAATGGACCACCTTCCTGGTCCACCGTTGAGCTTGATGGTGCCATTAATGACACCCTCTGTGTTAACTCTCGCCAGTTCACTGTACATCTTAACACTACAGTACACTATACCGCTGCACACTTGTTCGGCACGGAATTTGTTGCTGGCACGTGTCCATTCACATTACCAAACATTAACAATTACTTAACGTTTGGTTCCATATGTTTTTCCACTGTTAACAATGGTGGCTGCACTATACATGTTCAAAAAGTGTTGCGACACTATCGCTATACTTTTGGCACTATCTATGTTTCCTATCAACCAGGAAACCAAATAACAGCTATGCCAAAAGCCTCTTCAGGCACTACTGACATATCCACTGTTTATCTTAACGTGTGTACCAAGTATAATATCTATGGTAAAACTGGTACAGGCGTCATTACTAAAACAAATGAAACCCACATTGGAGGACTTTACTACTCCTCCCTTAGTGGTGACTTATTAGCATTTAAAAACTCAACTACACAAACAATTTACAGCATCACACCCTGTGAATTGTCTGCACAAGTTGCTGTATATAATGATTCTATTATTGCAGCATTCACTTCAACAGAGAATTTCACCTTCTCTGATTTCTCTTATAAGCTGAAAACTCCCATGTTCTATTACCACTCAATTGGTAATACGACATGTGAAGCACCAGCTATTACATTTGGTTCTATTGGTGTCTGCCCAGATGGTGGCCTAATTATTCAAAGTGCCACCACCAATGATGTAGACGCTGTAGTGCCTATTTCCACACAGAATATCTCAATACCTATCAACTTCACTGTATCTATTCAGACTGAGTACATACAGATTGAGCACCATCCTATTACGGTGGATTGCAGAAAATATGTTTGCAATGGCAACCCTCGTTGCCTGCAACTACTACTACAATATACTAGTGCGTGTTCAACAATTGAGCAGGCTCTAGCTTTAAATGCGCGACTTGAAGCAGCTAGCATCCAGTCAATGCTCACATACAACCCTCAGACAGTCAAGCTTGCAAACATTACAAACTTTCAGTCTGATGGTATCAAATATGACTTATCCTCCATACTACCTGTGTCAACTGGATCCAGGTCTGCTATTGAGGATTTACTCTTTGATAAAGTTGTCACAAATGGTCTAGGAACAGTAGACCAAGACTATAAAAAGTGTACCAACGGTTATTCCATTGCTGACCTTGTTTGTGCACAATATTATAATGGTATTATGGTACTTCCAGGCGTCGCTGACCCTGAGAAACTAGCCCAGTACACCGCTTCACTTACAGGTGCTATGGTCTTCGGTGGACTCACATCTGCCGCAGCCATACCATTTTCTCTAGCAGTCCAATCGCGTTTGAATTATGTTGCTCTTCAAACGGATGTGCTGCAACGCAACCAACAAATCTTGGCTACATCATTCAATAATGCTATGGGCAATATTACCCAGGCATTCCATGATGTCAACCAAGGACTTTCACAAGTTGCAGGTGCTGTCACCACGATAGCCAATGCATTCACAAAAATTCAGGATGTTGTCAATGCACAGGGCACTGCTCTCTCAACTCTGACAACACAGTTAACAAACAACTTTCAGGCTATCTCAGCCTCTATTGCTGACATTTATAATCGTCTCAACCAAATTGAGGCAGATGCACAAGTAGACAGACTCATTACAGGCAGGTTAGCTGCTCTCAATGCGTTTGTCACACAAACATTAGCTAAACTTGCTGAGGTTAGACAATCTCGCCAGTTAGCTCTTGACAAAGTTAATGAGTGTGTTAAATCGCAATCCGCTCGTTACGGATTTTGCGGCAACGGAACTCATCTTTTCTCATTTTCAAATGCAGCACCGTATGGACTGATGTTTTTCCACACTGTTCTGCTTCCCACACAATACGCCACAGTGCAAGCCTATTCAGGTATTTGCCACCAGAATAGAGCACTTACACTGAGAGACCCATCACTTGCACTCTTCCAGAAAGATGACAAGTATTTAATCACACCACGCAATATGTACCAACCCCGAACTGCCACCAAGGCGGACTTCGTGTACATACAGTCGTGTGACATCACATTCCTCAATCTAACTGACACTACTATTGAAGCCGTCATACCTGATTATGTCGACGTCAATAAAACAATTGAGGAGTTTCTCAACAACTTACCTAACTATACACTGCCAGATCTTTCGATCGACAGGTATAACAACACAATATTAAATCTCACCACTGAGATTGCTGATTTAAATGGTAAAGCTGCCAACCTCTCTCAAATTGTTGAGGAACTTGAGCAGTACATCAAAAACATTAACAGCACACTAGTTGACCTCGAATGGCTCAACCGTGTTGAAACCTACATCAAATGGCCATGGTATATATGGCTTGCCATAGCCCTGGCATTTACTGCATTTGTCGCAATCCTCATAACAATCTTTCTTTGCACTGGTTGCTGTGGAGGGTGCTTTGGTTGTTGCGGAGGTTGTTTTGGCCTTTTCTCAAAGAAGAAAAGGTATACAGACGACCATCCAACACCGTCCTTCAAATTTAAGGAATGGTAG
Protein
MQGALLITFIAVVTSSPLADKILDFLTFPEAHAYLFPQSRMVRNIEGGATERCMYVQEGGFIPDNFTFPQWFVLTNDSTLLQGELTTSQPLIVNGHLCSWQSNQQYRKYSFNTSCPLPADGCNPTWGSGNLKLFKGWNNTKGLTDNIRINVNISQSDYRTSAGSVGLELEGGGTVNITCTNSSTPVTTYSLLPWARSSGEPIYCFANVSSPSQSYIDFMGILPPFVSEIAFDRSGSIYINGYRYFKISPLLNVELQLNFNLTSDHFSVTWSNYTEVHLNTTNGYIHQIKYCYDPLDKLACEMNTFQLPDGVYPYTPAQQQALPETFVTTPVYANHTTVVVRTQYTVSSTGINGPPSWSTVELDGAINDTLCVNSRQFTVHLNTTVHYTAAHLFGTEFVAGTCPFTLPNINNYLTFGSICFSTVNNGGCTIHVQKVLRHYRYTFGTIYVSYQPGNQITAMPKASSGTTDISTVYLNVCTKYNIYGKTGTGVITKTNETHIGGLYYSSLSGDLLAFKNSTTQTIYSITPCELSAQVAVYNDSIIAAFTSTENFTFSDFSYKLKTPMFYYHSIGNTTCEAPAITFGSIGVCPDGGLIIQSATTNDVDAVVPISTQNISIPINFTVSIQTEYIQIEHHPITVDCRKYVCNGNPRCLQLLLQYTSACSTIEQALALNARLEAASIQSMLTYNPQTVKLANITNFQSDGIKYDLSSILPVSTGSRSAIEDLLFDKVVTNGLGTVDQDYKKCTNGYSIADLVCAQYYNGIMVLPGVADPEKLAQYTASLTGAMVFGGLTSAAAIPFSLAVQSRLNYVALQTDVLQRNQQILATSFNNAMGNITQAFHDVNQGLSQVAGAVTTIANAFTKIQDVVNAQGTALSTLTTQLTNNFQAISASIADIYNRLNQIEADAQVDRLITGRLAALNAFVTQTLAKLAEVRQSRQLALDKVNECVKSQSARYGFCGNGTHLFSFSNAAPYGLMFFHTVLLPTQYATVQAYSGICHQNRALTLRDPSLALFQKDDKYLITPRNMYQPRTATKADFVYIQSCDITFLNLTDTTIEAVIPDYVDVNKTIEEFLNNLPNYTLPDLSIDRYNNTILNLTTEIADLNGKAANLSQIVEELEQYIKNINSTLVDLEWLNRVETYIKWPWYIWLAIALAFTAFVAILITIFLCTGCCGGCFGCCGGCFGLFSKKKRYTDDHPTPSFKFKEW
Summary
Function
S1 region attaches the virion to the cell membrane by interacting with host ANPEP/aminopeptidase N, initiating the infection. Binding to the receptor probably induces conformational changes in the S glycoprotein unmasking the fusion peptide of S2 region and activating membranes fusion. S2 region belongs to the class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) regions assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer. During virus morphogenesis, found in a complex with M and HE proteins. Interacts with host ANPEP.
Miscellaneous
Bat coronavirus 512/2005 is highly similar to porcine epidemic diarrhea virus (PEDV).
Similarity
Belongs to the alphacoronaviruses spike protein family.
Keywords
Coiled coil
Glycoprotein
Host membrane
Host-virus interaction
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Virion
Virulence
Virus entry into host cell
3D-structure
Feature
chain Spike glycoprotein
Uniprot
H9BR00
H9BR08
A0A2Z5XXS4
A0A2Z5XXT6
A0A2Z5XXR5
A0A1L2KGB2
+ More
A0A0U1WJW2 A0A1L2KGD3 A0A1L2KGD0 F1DB14 A0A482K0W7 A0A1W5YKS6 A0A0U1UZ37 A0A3G3NGX8 A0A0P0KR98 A0A3G3NHH0 A0A3G3NGW9 A0A3G3NHL7 A0A0P0KBC5 A4ULL1 A0A482JZK4 K4JZU5 A0A0N7J128 A0A0P0K0X5 K4KCP3 A0A0P0KXK6 A0A0P0KI80 A0A4Y6GL52 A0A3G3NGT9 A0A0N7J129 A0A3G3NGQ0 A0A3G3NGS7 A0A1B3TNA4 A0A4Y6GL65 A0A3G3NGQ4 A0A4Y6GL57 A0A1L2KGE4 A0A3G3NGM2 A0A3G3NGM9 A0A3G3NGG7 A0A3G3NHH9 A0A1L2KGF1 A0A1W5YKT3 A0A219TUS3 A0A3G3NH05 A0A3G3NHI3 A0A4Y5QL66 A0A4Y6GL18 Q0Q466 Q0Q4F5 I1VWF5 A0A3G3NGN7 A0A3G3NGV3 Q0Q4F6 A0A0U2GQ76 A0A0U2GN50 A0A0U2GNG3 A0A0U2GMQ4 A0A0U2GRL6 A0A0U2GRB2 A0A3G3NGK2 A0A482JX42 A0A0U2GME6 A0A0U2GMN8 A0A3G3NHJ0 Q1HVM3 Q1HVL9 A0A0U2GMG2 A0A1C8YZ37 Q1HVM0 Q1HVM5 A0A1C8YZ26 A0A0U2GQI5 A0A059SGA9 Q1HVM1 A0A482JZV2 A0A0P0KRB3 A0A0P0KWD4 A0A3G3NGX2 A0A1C8YZ34 A0A0U2GN82 Q1HVL7 A0A0P0KH07 A0A3G3NGU0 Q1HVL8 P15423 A0A0U1WHD9 A0A1L7B934 A0A3G3NGU7 A0A384RJX8 A0A1V0BZ86 A0A0P0K6L9 A0A3G3NGJ7 S5YNL7 A0A0P0KR90 A0A384R9G0 H1AG30 Q1HVM6 H1AG29 A0A1L7B908
A0A0U1WJW2 A0A1L2KGD3 A0A1L2KGD0 F1DB14 A0A482K0W7 A0A1W5YKS6 A0A0U1UZ37 A0A3G3NGX8 A0A0P0KR98 A0A3G3NHH0 A0A3G3NGW9 A0A3G3NHL7 A0A0P0KBC5 A4ULL1 A0A482JZK4 K4JZU5 A0A0N7J128 A0A0P0K0X5 K4KCP3 A0A0P0KXK6 A0A0P0KI80 A0A4Y6GL52 A0A3G3NGT9 A0A0N7J129 A0A3G3NGQ0 A0A3G3NGS7 A0A1B3TNA4 A0A4Y6GL65 A0A3G3NGQ4 A0A4Y6GL57 A0A1L2KGE4 A0A3G3NGM2 A0A3G3NGM9 A0A3G3NGG7 A0A3G3NHH9 A0A1L2KGF1 A0A1W5YKT3 A0A219TUS3 A0A3G3NH05 A0A3G3NHI3 A0A4Y5QL66 A0A4Y6GL18 Q0Q466 Q0Q4F5 I1VWF5 A0A3G3NGN7 A0A3G3NGV3 Q0Q4F6 A0A0U2GQ76 A0A0U2GN50 A0A0U2GNG3 A0A0U2GMQ4 A0A0U2GRL6 A0A0U2GRB2 A0A3G3NGK2 A0A482JX42 A0A0U2GME6 A0A0U2GMN8 A0A3G3NHJ0 Q1HVM3 Q1HVL9 A0A0U2GMG2 A0A1C8YZ37 Q1HVM0 Q1HVM5 A0A1C8YZ26 A0A0U2GQI5 A0A059SGA9 Q1HVM1 A0A482JZV2 A0A0P0KRB3 A0A0P0KWD4 A0A3G3NGX2 A0A1C8YZ34 A0A0U2GN82 Q1HVL7 A0A0P0KH07 A0A3G3NGU0 Q1HVL8 P15423 A0A0U1WHD9 A0A1L7B934 A0A3G3NGU7 A0A384RJX8 A0A1V0BZ86 A0A0P0K6L9 A0A3G3NGJ7 S5YNL7 A0A0P0KR90 A0A384R9G0 H1AG30 Q1HVM6 H1AG29 A0A1L7B908
Pubmed
EMBL
JQ065045
AFD29209.1
JQ065046
AFD29217.1
LC364343
BBC54832.1
+ More
LC364344 BBC54842.1 LC364342 BBC54822.1 KY073744 APD51475.1 KJ473807 AIA62252.1 KY073745 APD51483.1 KY073746 APD51491.1 HQ728485 ADX59488.1 MG916904 QBP43290.1 KY770850 ARI44789.1 KJ473810 AIA62271.1 MH687963 AYR18567.1 KT253270 ALK28781.1 MH687946 AYR18473.1 MH687967 AYR18591.1 MH687964 AYR18573.1 KT253267 ALK28773.1 EF434381 ABO88151.1 MG916901 QBP43256.1 JQ989271 AFU92113.1 KT253261 KT253263 ALK28767.1 ALK28769.1 KT253265 ALK28771.1 JQ989270 AFU92104.1 KT253262 ALK28768.1 KT253266 ALK28772.1 MK211369 QDF43790.1 MH687962 AYR18561.1 KT253269 ALK28775.1 MH687949 AYR18487.1 MH687935 AYR18406.1 KU291449 AOG74789.1 MK211373 QDF43810.1 MH687939 AYR18430.1 MK211371 QDF43800.1 KY073747 APD51499.1 MH687934 AYR18400.1 MH687945 AYR18467.1 MH687936 MH687959 AYR18412.1 AYR18543.1 MH687950 AYR18493.1 KY073748 APD51507.1 KY770851 ARI44794.1 KT346372 ANV78630.1 MH687952 AYR18505.1 MH687938 AYR18424.1 MK720945 MK720946 QCX35167.1 QCX35178.1 MK211370 QDF43795.1 DQ648858 DQ648792 ABG11964.1 JQ410000 AFI49431.1 MH687951 AYR18499.1 MH687940 AYR18436.1 DQ648791 ABG11963.1 KT368899 ALA50137.1 KT368898 ALA50130.1 KT368916 ALA50256.1 KT368901 ALA50151.1 KT368903 ALA50165.1 KT368907 KT368908 ALA50193.1 ALA50200.1 MH687948 AYR18481.1 MG916902 QBP43268.1 KT368895 KT368896 MF593473 ALA50109.1 ALA50116.1 ATI09437.1 KT368892 KT368893 KT368894 KT368902 KT368904 KT368905 KT368906 KT368909 KT368910 KT368911 KT368912 KT368913 KT368914 ALA50088.1 ALA50095.1 ALA50102.1 ALA50158.1 ALA50172.1 ALA50179.1 ALA50186.1 ALA50207.1 ALA50214.1 ALA50221.1 ALA50228.1 ALA50235.1 ALA50242.1 MH687954 AYR18513.1 DQ243966 ABB90509.1 DQ243970 ABB90513.1 KT368897 ALA50123.1 KT253324 KT253326 KT253328 AOI28259.1 AOI28275.1 AOI28291.1 DQ243965 DQ243969 ABB90508.1 ABB90512.1 DQ243964 ABB90507.1 KT253325 AOI28267.1 KT368915 ALA50249.1 KF293664 KF293665 KF293666 AGW80948.1 DQ243967 DQ243968 ABB90510.1 ABB90511.1 MG916903 QBP43279.1 KT253272 ALK28793.1 KT253259 ALK28765.1 MH687965 AYR18579.1 KT253327 AOI28283.1 KT368900 ALA50144.1 DQ243972 ABB90515.1 KT253260 ALK28766.1 MH687960 AYR18549.1 DQ243971 ABB90514.1 X16816 AF304460 AF344186 AF344187 AF344188 AF344189 Y09923 Y10051 Y10052 X15654 KJ473800 AIA62234.1 KY369910 KY369911 APT69862.1 APT69869.1 MH687958 AYR18537.1 KM055532 KM055533 AIW52729.1 AIW52730.1 KY684760 ARA15429.1 KT253271 ALK28787.1 MH687941 AYR18442.1 KF514429 AGT21338.1 KT253264 ALK28770.1 KM055550 AIW52747.1 AB691764 BAL45638.1 DQ243963 ABB90506.1 AB691763 BAL45637.1 KY369909 KY674914 KY621348 APT69856.1 ARB07392.1 ARK08620.1
LC364344 BBC54842.1 LC364342 BBC54822.1 KY073744 APD51475.1 KJ473807 AIA62252.1 KY073745 APD51483.1 KY073746 APD51491.1 HQ728485 ADX59488.1 MG916904 QBP43290.1 KY770850 ARI44789.1 KJ473810 AIA62271.1 MH687963 AYR18567.1 KT253270 ALK28781.1 MH687946 AYR18473.1 MH687967 AYR18591.1 MH687964 AYR18573.1 KT253267 ALK28773.1 EF434381 ABO88151.1 MG916901 QBP43256.1 JQ989271 AFU92113.1 KT253261 KT253263 ALK28767.1 ALK28769.1 KT253265 ALK28771.1 JQ989270 AFU92104.1 KT253262 ALK28768.1 KT253266 ALK28772.1 MK211369 QDF43790.1 MH687962 AYR18561.1 KT253269 ALK28775.1 MH687949 AYR18487.1 MH687935 AYR18406.1 KU291449 AOG74789.1 MK211373 QDF43810.1 MH687939 AYR18430.1 MK211371 QDF43800.1 KY073747 APD51499.1 MH687934 AYR18400.1 MH687945 AYR18467.1 MH687936 MH687959 AYR18412.1 AYR18543.1 MH687950 AYR18493.1 KY073748 APD51507.1 KY770851 ARI44794.1 KT346372 ANV78630.1 MH687952 AYR18505.1 MH687938 AYR18424.1 MK720945 MK720946 QCX35167.1 QCX35178.1 MK211370 QDF43795.1 DQ648858 DQ648792 ABG11964.1 JQ410000 AFI49431.1 MH687951 AYR18499.1 MH687940 AYR18436.1 DQ648791 ABG11963.1 KT368899 ALA50137.1 KT368898 ALA50130.1 KT368916 ALA50256.1 KT368901 ALA50151.1 KT368903 ALA50165.1 KT368907 KT368908 ALA50193.1 ALA50200.1 MH687948 AYR18481.1 MG916902 QBP43268.1 KT368895 KT368896 MF593473 ALA50109.1 ALA50116.1 ATI09437.1 KT368892 KT368893 KT368894 KT368902 KT368904 KT368905 KT368906 KT368909 KT368910 KT368911 KT368912 KT368913 KT368914 ALA50088.1 ALA50095.1 ALA50102.1 ALA50158.1 ALA50172.1 ALA50179.1 ALA50186.1 ALA50207.1 ALA50214.1 ALA50221.1 ALA50228.1 ALA50235.1 ALA50242.1 MH687954 AYR18513.1 DQ243966 ABB90509.1 DQ243970 ABB90513.1 KT368897 ALA50123.1 KT253324 KT253326 KT253328 AOI28259.1 AOI28275.1 AOI28291.1 DQ243965 DQ243969 ABB90508.1 ABB90512.1 DQ243964 ABB90507.1 KT253325 AOI28267.1 KT368915 ALA50249.1 KF293664 KF293665 KF293666 AGW80948.1 DQ243967 DQ243968 ABB90510.1 ABB90511.1 MG916903 QBP43279.1 KT253272 ALK28793.1 KT253259 ALK28765.1 MH687965 AYR18579.1 KT253327 AOI28283.1 KT368900 ALA50144.1 DQ243972 ABB90515.1 KT253260 ALK28766.1 MH687960 AYR18549.1 DQ243971 ABB90514.1 X16816 AF304460 AF344186 AF344187 AF344188 AF344189 Y09923 Y10051 Y10052 X15654 KJ473800 AIA62234.1 KY369910 KY369911 APT69862.1 APT69869.1 MH687958 AYR18537.1 KM055532 KM055533 AIW52729.1 AIW52730.1 KY684760 ARA15429.1 KT253271 ALK28787.1 MH687941 AYR18442.1 KF514429 AGT21338.1 KT253264 ALK28770.1 KM055550 AIW52747.1 AB691764 BAL45638.1 DQ243963 ABB90506.1 AB691763 BAL45637.1 KY369909 KY674914 KY621348 APT69856.1 ARB07392.1 ARK08620.1
Proteomes
UP000142997
UP000162232
UP000204102
UP000106394
UP000277178
UP000144177
+ More
UP000122373 UP000162430 UP000155419 UP000118770 UP000113079 UP000146121 UP000136947 UP000116691 UP000152663 UP000173458 UP000103783 UP000120649 UP000151210 UP000101679 UP000169049 UP000100137 UP000103248 UP000108758 UP000110675 UP000115199 UP000115399 UP000124352 UP000128900 UP000136940 UP000152773 UP000153580 UP000171347 UP000173795 UP000147133 UP000153625 UP000130985 UP000104585 UP000006716 UP000126502 UP000096398
UP000122373 UP000162430 UP000155419 UP000118770 UP000113079 UP000146121 UP000136947 UP000116691 UP000152663 UP000173458 UP000103783 UP000120649 UP000151210 UP000101679 UP000169049 UP000100137 UP000103248 UP000108758 UP000110675 UP000115199 UP000115399 UP000124352 UP000128900 UP000136940 UP000152773 UP000153580 UP000171347 UP000173795 UP000147133 UP000153625 UP000130985 UP000104585 UP000006716 UP000126502 UP000096398
ProteinModelPortal
H9BR00
H9BR08
A0A2Z5XXS4
A0A2Z5XXT6
A0A2Z5XXR5
A0A1L2KGB2
+ More
A0A0U1WJW2 A0A1L2KGD3 A0A1L2KGD0 F1DB14 A0A482K0W7 A0A1W5YKS6 A0A0U1UZ37 A0A3G3NGX8 A0A0P0KR98 A0A3G3NHH0 A0A3G3NGW9 A0A3G3NHL7 A0A0P0KBC5 A4ULL1 A0A482JZK4 K4JZU5 A0A0N7J128 A0A0P0K0X5 K4KCP3 A0A0P0KXK6 A0A0P0KI80 A0A4Y6GL52 A0A3G3NGT9 A0A0N7J129 A0A3G3NGQ0 A0A3G3NGS7 A0A1B3TNA4 A0A4Y6GL65 A0A3G3NGQ4 A0A4Y6GL57 A0A1L2KGE4 A0A3G3NGM2 A0A3G3NGM9 A0A3G3NGG7 A0A3G3NHH9 A0A1L2KGF1 A0A1W5YKT3 A0A219TUS3 A0A3G3NH05 A0A3G3NHI3 A0A4Y5QL66 A0A4Y6GL18 Q0Q466 Q0Q4F5 I1VWF5 A0A3G3NGN7 A0A3G3NGV3 Q0Q4F6 A0A0U2GQ76 A0A0U2GN50 A0A0U2GNG3 A0A0U2GMQ4 A0A0U2GRL6 A0A0U2GRB2 A0A3G3NGK2 A0A482JX42 A0A0U2GME6 A0A0U2GMN8 A0A3G3NHJ0 Q1HVM3 Q1HVL9 A0A0U2GMG2 A0A1C8YZ37 Q1HVM0 Q1HVM5 A0A1C8YZ26 A0A0U2GQI5 A0A059SGA9 Q1HVM1 A0A482JZV2 A0A0P0KRB3 A0A0P0KWD4 A0A3G3NGX2 A0A1C8YZ34 A0A0U2GN82 Q1HVL7 A0A0P0KH07 A0A3G3NGU0 Q1HVL8 P15423 A0A0U1WHD9 A0A1L7B934 A0A3G3NGU7 A0A384RJX8 A0A1V0BZ86 A0A0P0K6L9 A0A3G3NGJ7 S5YNL7 A0A0P0KR90 A0A384R9G0 H1AG30 Q1HVM6 H1AG29 A0A1L7B908
A0A0U1WJW2 A0A1L2KGD3 A0A1L2KGD0 F1DB14 A0A482K0W7 A0A1W5YKS6 A0A0U1UZ37 A0A3G3NGX8 A0A0P0KR98 A0A3G3NHH0 A0A3G3NGW9 A0A3G3NHL7 A0A0P0KBC5 A4ULL1 A0A482JZK4 K4JZU5 A0A0N7J128 A0A0P0K0X5 K4KCP3 A0A0P0KXK6 A0A0P0KI80 A0A4Y6GL52 A0A3G3NGT9 A0A0N7J129 A0A3G3NGQ0 A0A3G3NGS7 A0A1B3TNA4 A0A4Y6GL65 A0A3G3NGQ4 A0A4Y6GL57 A0A1L2KGE4 A0A3G3NGM2 A0A3G3NGM9 A0A3G3NGG7 A0A3G3NHH9 A0A1L2KGF1 A0A1W5YKT3 A0A219TUS3 A0A3G3NH05 A0A3G3NHI3 A0A4Y5QL66 A0A4Y6GL18 Q0Q466 Q0Q4F5 I1VWF5 A0A3G3NGN7 A0A3G3NGV3 Q0Q4F6 A0A0U2GQ76 A0A0U2GN50 A0A0U2GNG3 A0A0U2GMQ4 A0A0U2GRL6 A0A0U2GRB2 A0A3G3NGK2 A0A482JX42 A0A0U2GME6 A0A0U2GMN8 A0A3G3NHJ0 Q1HVM3 Q1HVL9 A0A0U2GMG2 A0A1C8YZ37 Q1HVM0 Q1HVM5 A0A1C8YZ26 A0A0U2GQI5 A0A059SGA9 Q1HVM1 A0A482JZV2 A0A0P0KRB3 A0A0P0KWD4 A0A3G3NGX2 A0A1C8YZ34 A0A0U2GN82 Q1HVL7 A0A0P0KH07 A0A3G3NGU0 Q1HVL8 P15423 A0A0U1WHD9 A0A1L7B934 A0A3G3NGU7 A0A384RJX8 A0A1V0BZ86 A0A0P0K6L9 A0A3G3NGJ7 S5YNL7 A0A0P0KR90 A0A384R9G0 H1AG30 Q1HVM6 H1AG29 A0A1L7B908
PDB
6U7H
E-value=0,
Score=2360
Ontologies
GO
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
Subcellular Location
From MSLVP
Single-Pass Membrane
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Topology
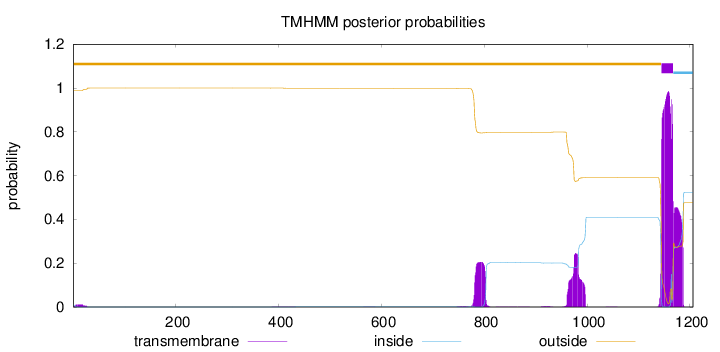
Length:
1206
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
40.65683
Exp number, first 60 AAs:
0.21607
Total prob of N-in:
0.01076
outside
1 - 1144
TMhelix
1145 - 1167
inside
1168 - 1206
Population Genetic Test Statistics
Pi
0.3231513
Theta
0.472782
Tajima's D
-0.9942683
CLR
8.572063
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 |
Protein |
YP_005352846.1 |
|
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.