Strain
CHINA_AVIAN_2007_NC_016994
(Region: China: Hong Kong; Strain: Night-heron coronavirus HKU19, complete genome.; Date: 2007)
Gene
membrane protein
Description
Annotated in NCBI,
membrane protein
Location
GenBank Accession
Full name
Membrane protein
Alternative Name
E1 glycoprotein
Matrix glycoprotein
Membrane glycoprotein
Matrix glycoprotein
Membrane glycoprotein
Sequence
CDS
ATGTCTGACGCTGCAGAGTGGCAACTCATCGTATTTATCATCTTAATCTGGGCCTTAGGCTTCATACTCCAAGGAGGGTATGCAGCAAGACATAAGGTCATTTATGTAATAAAACTAATACTGCTTTGGCTGCTCCAGCCTTTCACACTAGTAGTCACTATCTGGACTGCAGTAGACAATGGAGCCCAACCAAGCAGTGCAGTGTTTATTATTGCTATTATCTTCGCCATACTAACTTTCGTTATATGGCTGAAGTACTGGTATGACTCCATCAGACTTGTAATCAAAACAAAGTCAGGTTGGAGTTTCTCTCCAGAAACTAGACTACTTGTGTGTGCAATTGATGGAATGGGTAATGTCAAATGTGCACCAGTAGATCATTTACCAACAGCTCTCACTCCTGTCCTTGTAATGGGAAGGTTTATGCTAAACGGGCAGCTCCTTATGGCACAACAAACTGTGCAGACAGCTCCTAGAGCTTTGTATGTCATGACACCGAGTCAGACATATCATTTTACTTTGAAGAAAACTTTTCAAGATCCAGACTTCAAAGACACAGCCACATTTTGTTACCTTGTCGACCGCATTTCCAAGGCAGATTTGCAGAGTGTGACAACAGGCAGCAATTACGCACTGTACAAACACCTTTAA
Protein
MSDAAEWQLIVFIILIWALGFILQGGYAARHKVIYVIKLILLWLLQPFTLVVTIWTAVDNGAQPSSAVFIIAIIFAILTFVIWLKYWYDSIRLVIKTKSGWSFSPETRLLVCAIDGMGNVKCAPVDHLPTALTPVLVMGRFMLNGQLLMAQQTVQTAPRALYVMTPSQTYHFTLKKTFQDPDFKDTATFCYLVDRISKADLQSVTTGSNYALYKHL
Summary
Function
Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins.
Subunit
Homomultimer. Interacts with envelope E protein in the budding compartment of the host cell, which is located between endoplasmic reticulum and the Golgi complex. Forms a complex with HE and S proteins. Interacts with nucleocapsid N protein. This interaction probably participates in RNA packaging into the virus.
Similarity
Belongs to the gammacoronaviruses M protein family.
Belongs to the betacoronaviruses M protein family.
Belongs to the betacoronaviruses M protein family.
Keywords
Glycoprotein
Host Golgi apparatus
Host membrane
Host-virus interaction
Membrane
Transmembrane
Transmembrane helix
Viral envelope protein
Viral immunoevasion
Viral matrix protein
Virion
Feature
chain Membrane protein
Uniprot
H9BR19
A0A2U9I694
H9BR27
B6VDY9
A0A2Z5XXU6
A0A145YPB1
+ More
A0A140ES76 A0A0U3BGC8 B6VDY0 A0A2Z4EVY0 A0A1C9C4I5 A0A1Z4F680 X2EZT4 A0A075X0D1 A6N254 A0A075E3D6 A0A075E3P1 A0A075E3D0 A0A097EU50 W8R479 A0A075E3L3 H9BQY0 A0A0E3U220 A0A2H4NFS0 A0A286NGL7 A0A1Z4F693 H9BQY7 A0A5J6NBG5 A0A140ESG7 A0A514C923 H9BQZ4 A0A2Z5XXS3 H9BR10 A0A5B8JML2 A0A2Z5XXR4 A0A2Z5XXT2 A0A2Z4G602 A0A2Z4G622 A0A2Z4G609 B6VDX1 A0A4Y6ER58 B6VDW2 A0A2Z4EVV0 A0A2Z4EVU8 A0A346IMW4 H9BR02 F4MIR0 A0A2R4KSQ9 A0A516UWD7 Q91SA2 A0A1Z3FR08 A0A3S7GYH1 Q6W359 A0A1X9Y2Y2 A0A0C5B155 A0A1V0JAZ7 A0A166MX93 A0A346M350 A0A2P1IQA5 C9DST0 C6GHN0 A4ZU79 C6GHP2 A4ZU13 A4ZU68 C6GHQ3 A4ZTY0 B7U2P8 A4ZU46 A4ZTZ1 A4ZU35 Q06BD5 A4ZU57 A4ZU02 Q4U3R5 Q8V433 B7U2S0 B7U2N8 B7U2M7 C6GHL8 B7U2L6 A3FEW1 Q0PL34 Q4U3R7 A3FEW0 Q4U3R0 A0A191URC8 B7U2Q9 P69600 P69599 A4ZU24 A0A346PYP7 Q91A23 G4RJ87 B3VZS6 Q9J3N8 G4RJ86 G4RJ85 A0A0N9QUP2
A0A140ES76 A0A0U3BGC8 B6VDY0 A0A2Z4EVY0 A0A1C9C4I5 A0A1Z4F680 X2EZT4 A0A075X0D1 A6N254 A0A075E3D6 A0A075E3P1 A0A075E3D0 A0A097EU50 W8R479 A0A075E3L3 H9BQY0 A0A0E3U220 A0A2H4NFS0 A0A286NGL7 A0A1Z4F693 H9BQY7 A0A5J6NBG5 A0A140ESG7 A0A514C923 H9BQZ4 A0A2Z5XXS3 H9BR10 A0A5B8JML2 A0A2Z5XXR4 A0A2Z5XXT2 A0A2Z4G602 A0A2Z4G622 A0A2Z4G609 B6VDX1 A0A4Y6ER58 B6VDW2 A0A2Z4EVV0 A0A2Z4EVU8 A0A346IMW4 H9BR02 F4MIR0 A0A2R4KSQ9 A0A516UWD7 Q91SA2 A0A1Z3FR08 A0A3S7GYH1 Q6W359 A0A1X9Y2Y2 A0A0C5B155 A0A1V0JAZ7 A0A166MX93 A0A346M350 A0A2P1IQA5 C9DST0 C6GHN0 A4ZU79 C6GHP2 A4ZU13 A4ZU68 C6GHQ3 A4ZTY0 B7U2P8 A4ZU46 A4ZTZ1 A4ZU35 Q06BD5 A4ZU57 A4ZU02 Q4U3R5 Q8V433 B7U2S0 B7U2N8 B7U2M7 C6GHL8 B7U2L6 A3FEW1 Q0PL34 Q4U3R7 A3FEW0 Q4U3R0 A0A191URC8 B7U2Q9 P69600 P69599 A4ZU24 A0A346PYP7 Q91A23 G4RJ87 B3VZS6 Q9J3N8 G4RJ86 G4RJ85 A0A0N9QUP2
Pubmed
22278237
29875375
18971277
27231358
27718337
29872066
+ More
27424024 28634353 24723718 25759498 17459938 24723704 24964136 25153521 24744332 26584185 26337879 26514769 26250097 25817405 26982324 28758347 29621617 29695146 29569144 31239538 29363281 30758768 21539870 28495648 29508550 20022075 17344285 17434558 18842722 17459444 16604443 11714968 27274850 9778786 30126059 11087096 26262637
27424024 28634353 24723718 25759498 17459938 24723704 24964136 25153521 24744332 26584185 26337879 26514769 26250097 25817405 26982324 28758347 29621617 29695146 29569144 31239538 29363281 30758768 21539870 28495648 29508550 20022075 17344285 17434558 18842722 17459444 16604443 11714968 27274850 9778786 30126059 11087096 26262637
EMBL
JQ065047
AFD29228.1
MH013338
AWR88317.1
JQ065048
AFD29236.1
+ More
FJ376622 ACJ12064.1 LC364345 LC364346 BBC54854.1 BBC54863.1 KU984334 KU870479 KU870480 KU870481 KU870482 AMW88199.1 APC23093.1 APC23094.1 APC23095.1 APC23096.1 KR265851 AML40806.1 KT313678 KT313680 ALT00614.1 ALT00616.1 FJ376621 ACJ12055.1 MG812377 AWV67127.1 KX118627 AOM53029.1 LC216914 BAY00725.1 KJ567050 AHM88402.1 KJ769231 AIH06859.1 EF584908 ABQ39960.1 KJ601779 AIB07802.1 KJ601778 AIB07795.1 KJ601777 AIB07787.1 KM820765 AIT11902.1 KJ481931 AHL45009.1 KJ601780 AIB07809.1 JQ065042 KJ462462 KJ569769 KJ584355 KJ584356 KJ584357 KJ584358 KJ584359 KJ584360 KJ620016 KM012168 KT381613 AFD29189.1 AHN16222.1 AHN16229.1 AIA60975.1 AIA60982.1 AIA60989.1 AIA60996.1 AIA61003.1 AIA61009.1 AIA99520.1 AIP90488.1 ALJ32187.1 KP981395 KP757890 KP757891 KP757892 KT266822 KR131621 KT021234 KT336560 KT313675 KT313676 KT313677 KT313679 KT313681 KT313682 KT313683 KT313684 KT313685 KT313686 KR150443 KR265847 KR265848 KR265849 KR265850 KR265852 KR265853 KR265854 KR265855 KR265856 KR265857 KR265858 KR265859 KR265860 KR265861 KR265862 KR265863 KR265865 KU665558 KU051641 KU051642 KU051643 KU051644 KU051645 KU051646 KU051647 KU051648 KU051649 KU051650 KU051651 KU051652 KU051653 KU051654 KU051655 KU051656 KU517165 KU981059 KU981060 KU981061 KU981062 KX022602 KX022603 KX022604 KX022605 KU870483 KU870484 KY065120 KY129985 KX834351 KX834352 KX443143 KY354363 KY354364 KY364365 KY293677 KY293678 MF431742 MF431743 KY926512 MF948005 KY363868 KY513724 KY513725 MF041982 MF280390 MG242062 MG837130 MG837131 MG837132 MG837133 MH708123 MH708124 MH708125 MG832584 MH715491 LC260045 LC260038 LC260039 LC260040 LC260041 LC260042 LC260043 LC260044 MK005882 MK330605 AKA97940.1 AKC54430.1 AKC54437.1 AKC54444.1 AKQ63085.1 ALA13747.1 ALD83758.1 ALS54088.1 ALT00611.1 ALT00612.1 ALT00613.1 ALT00615.1 ALT00617.1 ALT00618.1 ALT00619.1 ALT00620.1 ALT00621.1 ALT00622.1 AML40606.1 AML40778.1 AML40785.1 AML40792.1 AML40799.1 AML40813.1 AML40820.1 AML40827.1 AML40834.1 AML40841.1 AML40848.1 AML40855.1 AML40862.1 AML40869.1 AML40876.1 AML40883.1 AML40890.1 AML40904.1 AML83918.1 AMN91623.1 AMN91629.1 AMN91635.1 AMN91641.1 AMN91647.1 AMN91653.1 AMN91659.1 AMN91665.1 AMN91672.1 AMN91678.1 AMN91684.1 AMN91690.1 AMN91696.1 AMN91702.1 AMN91708.1 AMN91714.1 ANA78298.1 ANA78445.1 ANI85827.1 ANI85834.1 ANI85841.1 ANI85848.1 APC23097.1 APC23098.1 APG38200.1 APW35736.1 APZ76692.1 APZ76700.1 AQS99155.1 AQT34132.1 AQT34138.1 ARJ31776.1 ASK86336.1 ASK86343.1 ASR75141.1 ASR75152.1 ASS83088.1 ATJ00131.1 ATV90800.1 AUG59158.1 AUG59166.1 AUH28252.1 AVR48521.1 AVZ61084.1 AWG96867.1 AWG96874.1 AWG96880.1 AWG96886.1 AXP32218.1 AXP32225.1 AXP32232.1 AYP31080.1 AYU65234.1 BAZ95611.1 BBA66307.1 BBA66314.1 BBA66321.1 BBA66328.1 BBA66335.1 BBA66342.1 BBA66349.1 QCO76965.1 QDP14520.1 KY363867 ATV90796.1 MF095123 KY398011 MK572803 MK330604 ASY98689.1 ATU88975.1 QDH76196.1 QDP14513.1 LC216915 BAY00732.1 JQ065043 AFD29196.1 MK673766 QEX51275.1 KR265864 AML40897.1 MK355396 QDH76190.1 JQ065044 AFD29203.1 LC364343 BBC54834.1 JQ065046 AFD29219.1 MK204388 QDY92323.1 LC364342 BBC54824.1 LC364344 BBC54844.1 MF642322 AWV96561.1 MF642325 AWV96582.1 MF642323 MF642324 AWV96568.1 AWV96575.1 FJ376620 ACJ12046.1 MK211169 QDF21513.1 FJ376619 ACJ12037.1 MG812375 MG812376 AWV67109.1 AWV67118.1 MG812378 AWV67136.1 MH532440 AXP20283.1 JQ065045 AFD29211.1 FJ904714 ADA83481.1 MF083115 AVV64338.1 MK581202 QDQ69125.1 AF363599 AAK50026.1 KY348417 KY994645 ASC42971.1 AUF40279.1 MG757138 MG757139 MG757140 MG757141 MG757142 AVI15005.1 AVI15016.1 AVI15027.1 AVI15038.1 AVI15049.1 AY316299 FJ415324 AAQ67201.1 ACJ35488.1 KY805846 ARS23156.1 KM985633 AJL33726.1 KX982264 ARD06004.1 KU127229 ANA52020.1 MG913342 AXQ05194.1 MG518518 AVN88327.1 GQ427173 ACV87238.1 FJ938064 ACT11000.1 EF424624 ABP38331.1 FJ938065 ACT11012.1 EF424618 ABP38270.1 EF424623 ABP38325.1 FJ938066 ACT11023.1 EF424615 ABP38238.1 FJ425187 ACJ66984.1 EF424621 ABP38305.1 EF424616 ABP38250.1 EF424620 ABP38285.1 DQ915164 ABI94002.1 EF424622 ABP38308.1 EF424617 ABP38255.1 DQ016129 DQ016130 DQ016131 DQ016132 DQ016134 AAY33933.1 AAY33936.1 AAY33939.1 AAY33942.1 AAY33948.1 AF391542 FJ425189 ACJ67005.1 FJ425186 ACJ66966.1 FJ425185 ACJ66958.1 FJ938063 ACT10988.1 FJ425184 ACJ66952.1 EF371516 ABN49621.1 DQ811784 ABG89282.1 AH014866 AAY33930.1 EF371515 ABN49620.1 AH014871 AH014873 KU886219 AAY33945.1 AAY33951.1 ANJ04979.1 KU558922 KU558923 ANJ04722.1 ANJ04733.1 FJ425188 FJ425190 ACJ66992.1 AF058944 AF058943 EF424619 ABP38278.1 MG021194 AXR85436.1 AF391541 FJ849812 ACZ67634.1 EU826086 ACE95722.1 AF202999 AF206266 AF206267 AF206268 AF206269 AF206270 AF206271 FJ849811 ACZ67633.1 FJ849810 ACZ67632.1 KR231009 ALH21115.1
FJ376622 ACJ12064.1 LC364345 LC364346 BBC54854.1 BBC54863.1 KU984334 KU870479 KU870480 KU870481 KU870482 AMW88199.1 APC23093.1 APC23094.1 APC23095.1 APC23096.1 KR265851 AML40806.1 KT313678 KT313680 ALT00614.1 ALT00616.1 FJ376621 ACJ12055.1 MG812377 AWV67127.1 KX118627 AOM53029.1 LC216914 BAY00725.1 KJ567050 AHM88402.1 KJ769231 AIH06859.1 EF584908 ABQ39960.1 KJ601779 AIB07802.1 KJ601778 AIB07795.1 KJ601777 AIB07787.1 KM820765 AIT11902.1 KJ481931 AHL45009.1 KJ601780 AIB07809.1 JQ065042 KJ462462 KJ569769 KJ584355 KJ584356 KJ584357 KJ584358 KJ584359 KJ584360 KJ620016 KM012168 KT381613 AFD29189.1 AHN16222.1 AHN16229.1 AIA60975.1 AIA60982.1 AIA60989.1 AIA60996.1 AIA61003.1 AIA61009.1 AIA99520.1 AIP90488.1 ALJ32187.1 KP981395 KP757890 KP757891 KP757892 KT266822 KR131621 KT021234 KT336560 KT313675 KT313676 KT313677 KT313679 KT313681 KT313682 KT313683 KT313684 KT313685 KT313686 KR150443 KR265847 KR265848 KR265849 KR265850 KR265852 KR265853 KR265854 KR265855 KR265856 KR265857 KR265858 KR265859 KR265860 KR265861 KR265862 KR265863 KR265865 KU665558 KU051641 KU051642 KU051643 KU051644 KU051645 KU051646 KU051647 KU051648 KU051649 KU051650 KU051651 KU051652 KU051653 KU051654 KU051655 KU051656 KU517165 KU981059 KU981060 KU981061 KU981062 KX022602 KX022603 KX022604 KX022605 KU870483 KU870484 KY065120 KY129985 KX834351 KX834352 KX443143 KY354363 KY354364 KY364365 KY293677 KY293678 MF431742 MF431743 KY926512 MF948005 KY363868 KY513724 KY513725 MF041982 MF280390 MG242062 MG837130 MG837131 MG837132 MG837133 MH708123 MH708124 MH708125 MG832584 MH715491 LC260045 LC260038 LC260039 LC260040 LC260041 LC260042 LC260043 LC260044 MK005882 MK330605 AKA97940.1 AKC54430.1 AKC54437.1 AKC54444.1 AKQ63085.1 ALA13747.1 ALD83758.1 ALS54088.1 ALT00611.1 ALT00612.1 ALT00613.1 ALT00615.1 ALT00617.1 ALT00618.1 ALT00619.1 ALT00620.1 ALT00621.1 ALT00622.1 AML40606.1 AML40778.1 AML40785.1 AML40792.1 AML40799.1 AML40813.1 AML40820.1 AML40827.1 AML40834.1 AML40841.1 AML40848.1 AML40855.1 AML40862.1 AML40869.1 AML40876.1 AML40883.1 AML40890.1 AML40904.1 AML83918.1 AMN91623.1 AMN91629.1 AMN91635.1 AMN91641.1 AMN91647.1 AMN91653.1 AMN91659.1 AMN91665.1 AMN91672.1 AMN91678.1 AMN91684.1 AMN91690.1 AMN91696.1 AMN91702.1 AMN91708.1 AMN91714.1 ANA78298.1 ANA78445.1 ANI85827.1 ANI85834.1 ANI85841.1 ANI85848.1 APC23097.1 APC23098.1 APG38200.1 APW35736.1 APZ76692.1 APZ76700.1 AQS99155.1 AQT34132.1 AQT34138.1 ARJ31776.1 ASK86336.1 ASK86343.1 ASR75141.1 ASR75152.1 ASS83088.1 ATJ00131.1 ATV90800.1 AUG59158.1 AUG59166.1 AUH28252.1 AVR48521.1 AVZ61084.1 AWG96867.1 AWG96874.1 AWG96880.1 AWG96886.1 AXP32218.1 AXP32225.1 AXP32232.1 AYP31080.1 AYU65234.1 BAZ95611.1 BBA66307.1 BBA66314.1 BBA66321.1 BBA66328.1 BBA66335.1 BBA66342.1 BBA66349.1 QCO76965.1 QDP14520.1 KY363867 ATV90796.1 MF095123 KY398011 MK572803 MK330604 ASY98689.1 ATU88975.1 QDH76196.1 QDP14513.1 LC216915 BAY00732.1 JQ065043 AFD29196.1 MK673766 QEX51275.1 KR265864 AML40897.1 MK355396 QDH76190.1 JQ065044 AFD29203.1 LC364343 BBC54834.1 JQ065046 AFD29219.1 MK204388 QDY92323.1 LC364342 BBC54824.1 LC364344 BBC54844.1 MF642322 AWV96561.1 MF642325 AWV96582.1 MF642323 MF642324 AWV96568.1 AWV96575.1 FJ376620 ACJ12046.1 MK211169 QDF21513.1 FJ376619 ACJ12037.1 MG812375 MG812376 AWV67109.1 AWV67118.1 MG812378 AWV67136.1 MH532440 AXP20283.1 JQ065045 AFD29211.1 FJ904714 ADA83481.1 MF083115 AVV64338.1 MK581202 QDQ69125.1 AF363599 AAK50026.1 KY348417 KY994645 ASC42971.1 AUF40279.1 MG757138 MG757139 MG757140 MG757141 MG757142 AVI15005.1 AVI15016.1 AVI15027.1 AVI15038.1 AVI15049.1 AY316299 FJ415324 AAQ67201.1 ACJ35488.1 KY805846 ARS23156.1 KM985633 AJL33726.1 KX982264 ARD06004.1 KU127229 ANA52020.1 MG913342 AXQ05194.1 MG518518 AVN88327.1 GQ427173 ACV87238.1 FJ938064 ACT11000.1 EF424624 ABP38331.1 FJ938065 ACT11012.1 EF424618 ABP38270.1 EF424623 ABP38325.1 FJ938066 ACT11023.1 EF424615 ABP38238.1 FJ425187 ACJ66984.1 EF424621 ABP38305.1 EF424616 ABP38250.1 EF424620 ABP38285.1 DQ915164 ABI94002.1 EF424622 ABP38308.1 EF424617 ABP38255.1 DQ016129 DQ016130 DQ016131 DQ016132 DQ016134 AAY33933.1 AAY33936.1 AAY33939.1 AAY33942.1 AAY33948.1 AF391542 FJ425189 ACJ67005.1 FJ425186 ACJ66966.1 FJ425185 ACJ66958.1 FJ938063 ACT10988.1 FJ425184 ACJ66952.1 EF371516 ABN49621.1 DQ811784 ABG89282.1 AH014866 AAY33930.1 EF371515 ABN49620.1 AH014871 AH014873 KU886219 AAY33945.1 AAY33951.1 ANJ04979.1 KU558922 KU558923 ANJ04722.1 ANJ04733.1 FJ425188 FJ425190 ACJ66992.1 AF058944 AF058943 EF424619 ABP38278.1 MG021194 AXR85436.1 AF391541 FJ849812 ACZ67634.1 EU826086 ACE95722.1 AF202999 AF206266 AF206267 AF206268 AF206269 AF206270 AF206271 FJ849811 ACZ67633.1 FJ849810 ACZ67632.1 KR231009 ALH21115.1
Proteomes
UP000132721
UP000167590
UP000103579
UP000112854
UP000171091
UP000011786
+ More
UP000126862 UP000139576 UP000104403 UP000129252 UP000160089 UP000114014 UP000102644 UP000172095 UP000103266 UP000118543 UP000122701 UP000124223 UP000147537 UP000149778 UP000152486 UP000152859 UP000156755 UP000161942 UP000174689 UP000097413 UP000100106 UP000108307 UP000115701 UP000116882 UP000117737 UP000118503 UP000120336 UP000120804 UP000130144 UP000131750 UP000132517 UP000136494 UP000137853 UP000141273 UP000144892 UP000147360 UP000148151 UP000150502 UP000151351 UP000154045 UP000156468 UP000158884 UP000161041 UP000162784 UP000163421 UP000169372 UP000170248 UP000172024 UP000180351 UP000180352 UP000103330 UP000118624 UP000135406 UP000162232 UP000116644 UP000161619 UP000142997 UP000133571 UP000158296 UP000123786 UP000136745 UP000147178 UP000139966 UP000173415 UP000163913 UP000162935 UP000121953 UP000158327 UP000136116 UP000127099 UP000170940 UP000154814 UP000139235 UP000151273 UP000008571 UP000133443 UP000140344 UP000104294 UP000172225 UP000174569 UP000147981 UP000102696 UP000148210 UP000134650 UP000008570 UP000097758
UP000126862 UP000139576 UP000104403 UP000129252 UP000160089 UP000114014 UP000102644 UP000172095 UP000103266 UP000118543 UP000122701 UP000124223 UP000147537 UP000149778 UP000152486 UP000152859 UP000156755 UP000161942 UP000174689 UP000097413 UP000100106 UP000108307 UP000115701 UP000116882 UP000117737 UP000118503 UP000120336 UP000120804 UP000130144 UP000131750 UP000132517 UP000136494 UP000137853 UP000141273 UP000144892 UP000147360 UP000148151 UP000150502 UP000151351 UP000154045 UP000156468 UP000158884 UP000161041 UP000162784 UP000163421 UP000169372 UP000170248 UP000172024 UP000180351 UP000180352 UP000103330 UP000118624 UP000135406 UP000162232 UP000116644 UP000161619 UP000142997 UP000133571 UP000158296 UP000123786 UP000136745 UP000147178 UP000139966 UP000173415 UP000163913 UP000162935 UP000121953 UP000158327 UP000136116 UP000127099 UP000170940 UP000154814 UP000139235 UP000151273 UP000008571 UP000133443 UP000140344 UP000104294 UP000172225 UP000174569 UP000147981 UP000102696 UP000148210 UP000134650 UP000008570 UP000097758
Pfam
PF01635
Corona_M
ProteinModelPortal
H9BR19
A0A2U9I694
H9BR27
B6VDY9
A0A2Z5XXU6
A0A145YPB1
+ More
A0A140ES76 A0A0U3BGC8 B6VDY0 A0A2Z4EVY0 A0A1C9C4I5 A0A1Z4F680 X2EZT4 A0A075X0D1 A6N254 A0A075E3D6 A0A075E3P1 A0A075E3D0 A0A097EU50 W8R479 A0A075E3L3 H9BQY0 A0A0E3U220 A0A2H4NFS0 A0A286NGL7 A0A1Z4F693 H9BQY7 A0A5J6NBG5 A0A140ESG7 A0A514C923 H9BQZ4 A0A2Z5XXS3 H9BR10 A0A5B8JML2 A0A2Z5XXR4 A0A2Z5XXT2 A0A2Z4G602 A0A2Z4G622 A0A2Z4G609 B6VDX1 A0A4Y6ER58 B6VDW2 A0A2Z4EVV0 A0A2Z4EVU8 A0A346IMW4 H9BR02 F4MIR0 A0A2R4KSQ9 A0A516UWD7 Q91SA2 A0A1Z3FR08 A0A3S7GYH1 Q6W359 A0A1X9Y2Y2 A0A0C5B155 A0A1V0JAZ7 A0A166MX93 A0A346M350 A0A2P1IQA5 C9DST0 C6GHN0 A4ZU79 C6GHP2 A4ZU13 A4ZU68 C6GHQ3 A4ZTY0 B7U2P8 A4ZU46 A4ZTZ1 A4ZU35 Q06BD5 A4ZU57 A4ZU02 Q4U3R5 Q8V433 B7U2S0 B7U2N8 B7U2M7 C6GHL8 B7U2L6 A3FEW1 Q0PL34 Q4U3R7 A3FEW0 Q4U3R0 A0A191URC8 B7U2Q9 P69600 P69599 A4ZU24 A0A346PYP7 Q91A23 G4RJ87 B3VZS6 Q9J3N8 G4RJ86 G4RJ85 A0A0N9QUP2
A0A140ES76 A0A0U3BGC8 B6VDY0 A0A2Z4EVY0 A0A1C9C4I5 A0A1Z4F680 X2EZT4 A0A075X0D1 A6N254 A0A075E3D6 A0A075E3P1 A0A075E3D0 A0A097EU50 W8R479 A0A075E3L3 H9BQY0 A0A0E3U220 A0A2H4NFS0 A0A286NGL7 A0A1Z4F693 H9BQY7 A0A5J6NBG5 A0A140ESG7 A0A514C923 H9BQZ4 A0A2Z5XXS3 H9BR10 A0A5B8JML2 A0A2Z5XXR4 A0A2Z5XXT2 A0A2Z4G602 A0A2Z4G622 A0A2Z4G609 B6VDX1 A0A4Y6ER58 B6VDW2 A0A2Z4EVV0 A0A2Z4EVU8 A0A346IMW4 H9BR02 F4MIR0 A0A2R4KSQ9 A0A516UWD7 Q91SA2 A0A1Z3FR08 A0A3S7GYH1 Q6W359 A0A1X9Y2Y2 A0A0C5B155 A0A1V0JAZ7 A0A166MX93 A0A346M350 A0A2P1IQA5 C9DST0 C6GHN0 A4ZU79 C6GHP2 A4ZU13 A4ZU68 C6GHQ3 A4ZTY0 B7U2P8 A4ZU46 A4ZTZ1 A4ZU35 Q06BD5 A4ZU57 A4ZU02 Q4U3R5 Q8V433 B7U2S0 B7U2N8 B7U2M7 C6GHL8 B7U2L6 A3FEW1 Q0PL34 Q4U3R7 A3FEW0 Q4U3R0 A0A191URC8 B7U2Q9 P69600 P69599 A4ZU24 A0A346PYP7 Q91A23 G4RJ87 B3VZS6 Q9J3N8 G4RJ86 G4RJ85 A0A0N9QUP2
PDB
3CH8
E-value=3.91226,
Score=66
Ontologies
GO
Subcellular Location
From MSLVP
Host nucleus > host nucleolus. Host cytoplasm. Secreted.
From Uniprot
Host Golgi apparatus membrane
Virion membrane
Virion membrane
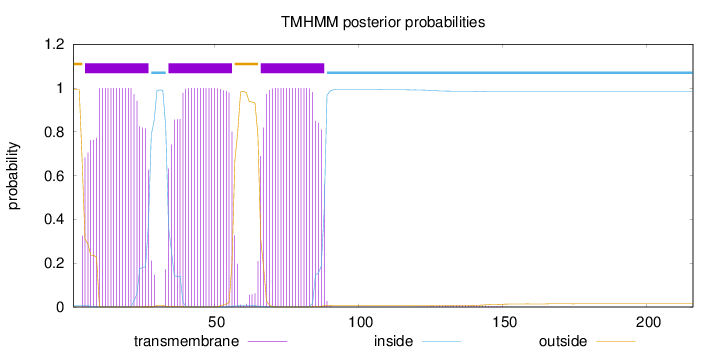
Topology
Length:
216
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
65.89283
Exp number, first 60 AAs:
43.75767
Total prob of N-in:
0.00737
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 27
inside
28 - 33
TMhelix
34 - 56
outside
57 - 65
TMhelix
66 - 88
inside
89 - 216
Population Genetic Test Statistics
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 |
Protein |
YP_005352865.1 |
|
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.