Strain
CHINA_AVIAN_2008_NC_016995
(Region: China: Hong Kong; Strain: Wigeon coronavirus HKU20, complete genome.; Date: 2008)
Gene
spike glycoprotein
Description
Annotated in NCBI,
spike glycoprotein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGTACAGGTTTGCTATACTAATGTTGGTCATTTCGCCAACCCTAGCTGCATCCATAGCTGATAAAGTTTTGGATGTAGTAACCTTTCCCGGAGCATCTCGGTTGTTGTACCCACAACACTACCAACAACCCCAAGTAAGGGGGTGGACTGATGATATTGGTGTTGATGGTCGCATACCTGCAACCTATCCACTAACCAACACCTTCTATCTTTCAAATACCACCACACCAGTTGATGGTGTTTATACTAGTCTTCAGCCCTTATTAATTCGTTGTAAGTTTAAATACGTCAATGATACTGTCAATGCTGGTAAAATTCTTACTATGTATTTTAACCTCACCAATAACACAGGTAGATGTGACGGGAATTCGGAAAAGGTTGGTCTTGTGGATGCCATCAGGTTTCAAATTAATGTCACAACCGACATATTACAGCTCACTGCAGGCAGTATTAACTTTACTACACCGGACGGAGAGCTGTATACATTACATTGTCTTAACACGACACTTGGTGTAGGCCAGGATTATTACCCATCTCAAAGCCGCACGCGGCAGTCTGGTGTTACCTATCATTGTTATATGATATATCGTAACACATCCTACGCCTACAGCGCTAATGAATACACTTATCGTGTGCTTCAATATTTGGGTCCCTTACCAGCATCAGTTAGGGAAATTGTTGCTTTTAGTAATGGATTCATTTACATTAATGGCATTCTTCTTTCGCGAATACCCGCGTTGCATAAAGTTGATTTTGGTCTTGTTAAGAGTGCCCATACGTCAGATTATTATGTTATCTTGTTTGCTGATATGGTTGACGTAATGGTCAACATATCCGCCACTGAAATGCAGTCTATCTTTTACTGCATAACCCCTTTTGAGCAGGTTAAATGTTCACAGAAACAAACCACTCTTTCTGATGGGTTCTATTCCACATCTGCCATTGAAGTCCTACAGCGCCAGCGCACATTTGTAGGGTTACCGATGGCTATTAACATTACTACGCTTAATTTTGCTATTAGTTATTTGAATACCAAGGCTAATCCTAGTTTAGGTGCACCACAGGAAGTCAATCTTACCATAAATGGGTTTGCAGATCAATTCTGTGTTACAACCTCCCAGTTTATGGTTACTCTTAATGTGACCTGTCTCGTTCATAATAATACTAGTATAATACCCTGTCCACAGGCTTACACGGCAGAAATCTCACCGGGGGATTGCCCATTTAATTTTTTGGATGTTAATAACTATTTAATGTTTGACAGCATTTGCTTTTCACCTTCTCCCAGTGGTGACAGCTGTCAGATGGTTATTAATAAAGTCTGGGCCAATAATCGCATACCTATGAGTAGCGTTTATGTCTCCTATGTCTATGGCAACCAGATAGTCGGTGTACCTCGACACGATCTCGTGCAGTTGGATCATGTCGTGTTTAATTTGTGCACAGACTACACCATTTATGGGTATACCGGTACTGGTGTTATACGTGAAACTAACACCACATACTTTGCCGGTTATGCCTATGCATCACCAGCTGGTCAACTCGTTGCCTATAAAAATCTTACTACTGGTGCTATTAATTCTATAACACCATGTAAGTTTAGTCAACAATTAGCCGTGTATAATGATACACCTGTAGCCCTTGTGTCAGCGGTGCCCTCGCAGGACTTTGGGTTTACCAATGCAACCTCTATGGGTACCTTTGTTGTCAATTCAAATGCAACTAATTGCACCATTCCAGCTCTTACTTATGGCCCACTTGGCATTTGTGCCAATGGCGCTCCATATGTAGTACCTACAGGTGAAGGCCAAGAACCTAGTGTTGTCCCCATTTCTACGGGCAACATTTCAATACCTGTCAATTTTACTGTAGCCATACAGCCTGAGTATCTACAGGTTTATACTGAGCAGGTTGTTGTAGACTGTGCCACTTATGTTTGCAACGGTAACCCATTGTGTAACAAGTTGTTGTTACAATACACTTCAGCTTGCGACACAATTGAGCAGGCGTTGCAAATGAGCGCCCGTTTAGAGTCTAGTGAAGTCTCTAACATGCTACACATTTCAACGCAGGCTCTGGATTTGGCTAACATCTCAAATTTTGAGGGTTATAATATGTCCTTTGTTTTGCCTGCTGTTAAACAGGGTAGGTCTGCCTTAGAAGACTTGCTATTTACTAAAGTAGTTACAGCTGGACTGGGCACAGTAGATGCTGACTATGAGAAATGTGCTAAAGGCATGGACATAGCAGACCTTGTATGTGTCCAGTACTATAACGGTATCATGGTGCTCCCCGGCGTAGCAAATGCTGGCAAAATGGCTGAGTACACAGCATCCCTTACTGGCGGTATGGTTATGGGTGGTATTACATCAGCAGCCTCTATACCATTTTCCTTAGCGGTACAAAGCCGCCTTAATTATGTAGCTCTGCAAACAGGTGTTATGTTAGATAATCAGAAGCTTCTAGCAGATTCCTTTAATAAGGCTATGGAGACAATTTCTGGTGCATTTAATTCACTTAATTCTGCTGTGCATGAAACTATTAATGTTGTTAACACTCTATCATCTGCACTTACTAAGATACAGAGTGTTGTCAATCAGCATACTACAGCCTTAAATCAGCTAACTCAGCAGTTGGCTAATAATTTCCAGGCTATATCTTCGTCTATTACAGACATTTATAACAGGTTGGGTCAACTAGAGGCTGATGCTCAGGTAGATCGGCTTATTACAGGTCGGTTGGCAGCCCTCAATGCATTTGTGTCCCAAACCTTAACCAAGGCAGAGCAGGTCCGCCAATCTAGGTTGCTTGCTCAGCAGAAAGTTAATGAGTGTGTCAAATCGCAGTCTCAGCGGTTTGGTTTCTGTGGCAATGGTTCTCACATCTTTACTATTGCCAATGCCGCGCCTGATGGCATTATGTTTTTGCACACGGTACTACAACCTGTAAGCTACATTACTGTTGTAGCCTATTCAGGGCTGTGTGTAGACAACACTTATGGCTACGCTCTTAAAGAGCCTAACCTAGCACTTGTGCAGTATAATGGTTATTATGTTACTCCGCGTAATATGTATCAACCACGTCCAGCTACTACAGCTGACTTTGTGCAAATACAATCTTGTGATGTTACCCTCTATAACATTACTTACAATAATATATCTTTGGTCATACCAGACTTTGTGGATACCAACAAAACTATTGAAGATATCATTTCTTCAATACCTAATAATACTGCGCCTAGTTTAGAAATAGCCAGTCTTAATCTGACTGTACTAAACCTGACGTCAGAGCTAGAACAATTAAAAGCCTCCACTGGCAATTTTACTCAAATTTCTCAGGAAATACAGGATTACATAAATAAACTCAATGACACTCTAGTTGACTTAGAGTGGCTCAACAGAGTAGAAACTTATGTTAAATGGCCATGGTGGGTGTGGCTTCTTATCTTTTTAGCCATTTGTACTTTTATAATAATTGTCGTAACAATCTTCCTATGCACAGGTTGCTGTGGTGGATGTTTTGGCTGTTTTGGTGGGTGCTGTGGTTTGTTTAGCAGAGTTAAACACCATGAGTTTTCGCACTTACCTGTAGAAGAACAAGATGGCTCAATACCAATTACCTATAAGAAAGTTAATTAA
Protein
MYRFAILMLVISPTLAASIADKVLDVVTFPGASRLLYPQHYQQPQVRGWTDDIGVDGRIPATYPLTNTFYLSNTTTPVDGVYTSLQPLLIRCKFKYVNDTVNAGKILTMYFNLTNNTGRCDGNSEKVGLVDAIRFQINVTTDILQLTAGSINFTTPDGELYTLHCLNTTLGVGQDYYPSQSRTRQSGVTYHCYMIYRNTSYAYSANEYTYRVLQYLGPLPASVREIVAFSNGFIYINGILLSRIPALHKVDFGLVKSAHTSDYYVILFADMVDVMVNISATEMQSIFYCITPFEQVKCSQKQTTLSDGFYSTSAIEVLQRQRTFVGLPMAINITTLNFAISYLNTKANPSLGAPQEVNLTINGFADQFCVTTSQFMVTLNVTCLVHNNTSIIPCPQAYTAEISPGDCPFNFLDVNNYLMFDSICFSPSPSGDSCQMVINKVWANNRIPMSSVYVSYVYGNQIVGVPRHDLVQLDHVVFNLCTDYTIYGYTGTGVIRETNTTYFAGYAYASPAGQLVAYKNLTTGAINSITPCKFSQQLAVYNDTPVALVSAVPSQDFGFTNATSMGTFVVNSNATNCTIPALTYGPLGICANGAPYVVPTGEGQEPSVVPISTGNISIPVNFTVAIQPEYLQVYTEQVVVDCATYVCNGNPLCNKLLLQYTSACDTIEQALQMSARLESSEVSNMLHISTQALDLANISNFEGYNMSFVLPAVKQGRSALEDLLFTKVVTAGLGTVDADYEKCAKGMDIADLVCVQYYNGIMVLPGVANAGKMAEYTASLTGGMVMGGITSAASIPFSLAVQSRLNYVALQTGVMLDNQKLLADSFNKAMETISGAFNSLNSAVHETINVVNTLSSALTKIQSVVNQHTTALNQLTQQLANNFQAISSSITDIYNRLGQLEADAQVDRLITGRLAALNAFVSQTLTKAEQVRQSRLLAQQKVNECVKSQSQRFGFCGNGSHIFTIANAAPDGIMFLHTVLQPVSYITVVAYSGLCVDNTYGYALKEPNLALVQYNGYYVTPRNMYQPRPATTADFVQIQSCDVTLYNITYNNISLVIPDFVDTNKTIEDIISSIPNNTAPSLEIASLNLTVLNLTSELEQLKASTGNFTQISQEIQDYINKLNDTLVDLEWLNRVETYVKWPWWVWLLIFLAICTFIIIVVTIFLCTGCCGGCFGCFGGCCGLFSRVKHHEFSHLPVEEQDGSIPITYKKVN
Summary
Function
S1 region attaches the virion to the cell membrane by interacting with host ANPEP/aminopeptidase N, initiating the infection. Binding to the receptor probably induces conformational changes in the S glycoprotein unmasking the fusion peptide of S2 region and activating membranes fusion. S2 region belongs to the class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) regions assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer. During virus morphogenesis, found in a complex with M and HE proteins. Interacts with host ANPEP.
Similarity
Belongs to the alphacoronaviruses spike protein family.
Keywords
3D-structure
Coiled coil
Glycoprotein
Host membrane
Host-virus interaction
Membrane
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Virion
Virulence
Virus entry into host cell
Reference proteome
Feature
chain Spike glycoprotein
Uniprot
H9BR25
A0A2U9I6C2
A0A482JX42
A0A482JZV2
A0A0U1UZ37
H9BR08
+ More
A0A1L2KGE4 A4ULL1 A0A0U1WJW2 A0A2Z5XXT6 A0A2Z5XXS4 A0A1L2KGB2 H9BR00 F1DB14 A0A1L2KGF1 A0A2Z5XXR5 A0A0P0KBC5 A0A0P0K0X5 A0A0N7J128 A0A0P0KXK6 A0A5B9BH95 A0A0P0G321 K4P2Y7 Q6Q1S2 A0A384RHS8 A0A384QZE9 A0A5B9BIJ4 A0A5B9MUE4 I7CMD1 A0A384RHT2 A0A384QXZ7 A0A1V0PKX8 A0A1V0E0U5 H9EJ44 A0A0K1D6F6 A0A1V0E0W0 H9EJV2 A0A0P0KR98 U3M7G5 Q06Y16 A0A384RBV6 A0A5B9BH24 A0A1L2YVI8 A0A2U8JDM5 H9EJ93 A0A384RK30 U3M6U5 H9EJA0 H9EJ51 U3M6Q4 H9EJ72 H9EJW4 A0A1V0PKW6 H9EJ86 A0A1V0PKX1 A0A0U2GNG3 A0A384R2Q9 U3M7H2 A0A513ZT30 A0A2H4ZBM8 U3M6S0 H9EJ37 A0A2L0WQE1 A0A0U2GQI5 A0A0U2GQ76 A0A0U2GMQ4 A0A0U2GRB2 A0A1C8YZ34 A0A0U2GN50 A0A0U2GRL6 A0A1B3TNA4 A0A0U2GMN8 A0A4Y5QL66 A0A1C8YZ37 A0A5B9BHU7 I1VWF5 H9EJ16 A0A1Y0EV12 A0A1C8YZ26 Q06Y10 A0A2H4ZBN0 A0A0U2GME6 P15423 A0A0U2GN82 A0A0U2GMG2 A0A0P0KI80 A0A059SGA9 Q1HVM6 H1AG29 A0A0N7J129 A0A5B9MSJ9 A0A0P0K6L9 A0A0P0KR90 A0A5B9BGI8 A0A0P0KMM1 Q1HVL3 Q1HVL0 S5YGU2 Q1HVL4 S5YAI3
A0A1L2KGE4 A4ULL1 A0A0U1WJW2 A0A2Z5XXT6 A0A2Z5XXS4 A0A1L2KGB2 H9BR00 F1DB14 A0A1L2KGF1 A0A2Z5XXR5 A0A0P0KBC5 A0A0P0K0X5 A0A0N7J128 A0A0P0KXK6 A0A5B9BH95 A0A0P0G321 K4P2Y7 Q6Q1S2 A0A384RHS8 A0A384QZE9 A0A5B9BIJ4 A0A5B9MUE4 I7CMD1 A0A384RHT2 A0A384QXZ7 A0A1V0PKX8 A0A1V0E0U5 H9EJ44 A0A0K1D6F6 A0A1V0E0W0 H9EJV2 A0A0P0KR98 U3M7G5 Q06Y16 A0A384RBV6 A0A5B9BH24 A0A1L2YVI8 A0A2U8JDM5 H9EJ93 A0A384RK30 U3M6U5 H9EJA0 H9EJ51 U3M6Q4 H9EJ72 H9EJW4 A0A1V0PKW6 H9EJ86 A0A1V0PKX1 A0A0U2GNG3 A0A384R2Q9 U3M7H2 A0A513ZT30 A0A2H4ZBM8 U3M6S0 H9EJ37 A0A2L0WQE1 A0A0U2GQI5 A0A0U2GQ76 A0A0U2GMQ4 A0A0U2GRB2 A0A1C8YZ34 A0A0U2GN50 A0A0U2GRL6 A0A1B3TNA4 A0A0U2GMN8 A0A4Y5QL66 A0A1C8YZ37 A0A5B9BHU7 I1VWF5 H9EJ16 A0A1Y0EV12 A0A1C8YZ26 Q06Y10 A0A2H4ZBN0 A0A0U2GME6 P15423 A0A0U2GN82 A0A0U2GMG2 A0A0P0KI80 A0A059SGA9 Q1HVM6 H1AG29 A0A0N7J129 A0A5B9MSJ9 A0A0P0K6L9 A0A0P0KR90 A0A5B9BGI8 A0A0P0KMM1 Q1HVL3 Q1HVL0 S5YGU2 Q1HVL4 S5YAI3
Pubmed
EMBL
JQ065048
AFD29234.1
MH013337
AWR88316.1
MG916902
QBP43268.1
+ More
MG916903 QBP43279.1 KJ473810 AIA62271.1 JQ065046 AFD29217.1 KY073747 APD51499.1 EF434381 ABO88151.1 KJ473807 AIA62252.1 LC364344 BBC54842.1 LC364343 BBC54832.1 KY073744 APD51475.1 JQ065045 AFD29209.1 HQ728485 ADX59488.1 KY073748 APD51507.1 LC364342 BBC54822.1 KT253267 ALK28773.1 KT253265 ALK28771.1 KT253261 KT253263 ALK28767.1 ALK28769.1 KT253262 ALK28768.1 MK334044 QED88019.1 KT381875 KU521535 ALJ53431.1 ANU06084.1 JX504050 AFV53148.1 AY567487 AY518894 AY758297 AY758298 AY758300 AY758301 KM055634 AIW52831.1 KM055642 AIW52839.1 MK334043 QED88012.1 MN306040 QEG03748.1 JX104161 JX524171 KM055632 KM055633 KM055635 KM055636 KM055637 KM055648 KM055649 KM055650 MG428699 MG428700 MG428701 MG428702 MG428703 MG428704 MG428705 MG428706 AFO70497.1 AFQ55884.1 AIW52829.1 AIW52830.1 AIW52832.1 AIW52833.1 AIW52834.1 AIW52845.1 AIW52846.1 AIW52847.1 AWK59931.1 AWK59937.1 AWK59943.1 AWK59949.1 AWK59955.1 AWK59961.1 AWK59967.1 AWK59973.1 KM055644 AIW52841.1 KM055639 KM055640 KM055641 KM055645 KM055647 AIW52836.1 AIW52837.1 AIW52838.1 AIW52842.1 AIW52844.1 KY554970 ARE29979.1 KY674916 ARB07406.1 JQ765567 AFD98778.1 KT266906 KX179494 KX179495 KX179496 AKT07952.1 APF29061.1 APF29062.1 APF29063.1 KY674915 KY554967 ARB07399.1 ARE29961.1 JQ771055 JQ771057 JQ771058 AFD64748.1 AFD64754.1 AFD64757.1 KT253270 ALK28781.1 KF530107 AGT51345.1 DQ445911 ABE97137.1 KM055638 AIW52835.1 MK334045 QED88026.1 KX179500 APF29071.1 MG428707 AWK59979.1 JQ765574 AFD98827.1 KM055643 AIW52840.1 KF530104 KF530106 KF530108 KF530109 KF530111 KF530113 KF530114 AGT51324.1 AGT51338.1 AGT51352.1 AGT51359.1 AGT51373.1 AGT51387.1 AGT51394.1 JQ765575 AFD98834.1 JQ765568 AFD98785.1 KF530105 AGT51331.1 JQ765571 JQ765572 AFD98806.1 AFD98813.1 JQ771059 AFD64760.1 KY554968 ARE29967.1 JQ765573 AFD98820.1 KY554969 KY554971 KY829118 ARE29973.1 ARE29985.1 ARK08627.1 KT368916 ALA50256.1 KM055646 AIW52843.1 KF530112 AGT51380.1 MN026166 QDH43736.1 MF996621 MF996622 AUF73167.1 KF530110 AGT51366.1 JQ765566 AFD98771.1 MG772808 AVA26873.1 KT368915 ALA50249.1 KT368899 ALA50137.1 KT368901 ALA50151.1 KT368907 KT368908 ALA50193.1 ALA50200.1 KT253327 AOI28283.1 KT368898 ALA50130.1 KT368903 ALA50165.1 KU291449 AOG74789.1 KT368892 KT368893 KT368894 KT368902 KT368904 KT368905 KT368906 KT368909 KT368910 KT368911 KT368912 KT368913 KT368914 ALA50088.1 ALA50095.1 ALA50102.1 ALA50158.1 ALA50172.1 ALA50179.1 ALA50186.1 ALA50207.1 ALA50214.1 ALA50221.1 ALA50228.1 ALA50235.1 ALA50242.1 MK720945 MK720946 QCX35167.1 QCX35178.1 KT253324 KT253326 KT253328 AOI28259.1 AOI28275.1 AOI28291.1 MK334046 QED88033.1 JQ410000 AFI49431.1 JQ765563 AFD98750.1 KY983586 ARU07594.1 KT253325 AOI28267.1 DQ445912 JQ771056 JQ771060 JQ765564 JQ765565 JQ765569 JQ765570 ABE97130.1 AFD64751.1 AFD64763.1 AFD98757.1 AFD98764.1 AFD98792.1 AFD98799.1 MF996623 AUF73168.1 KT368895 KT368896 MF593473 ALA50109.1 ALA50116.1 ATI09437.1 X16816 AF304460 AF344186 AF344187 AF344188 AF344189 Y09923 Y10051 Y10052 X15654 KT368900 ALA50144.1 KT368897 ALA50123.1 KT253266 ALK28772.1 KF293664 KF293665 KF293666 AGW80948.1 DQ243963 ABB90506.1 AB691763 BAL45637.1 KT253269 ALK28775.1 MN306018 QEG03731.1 KT253271 ALK28787.1 KT253264 ALK28770.1 MK334047 QED88040.1 KT253268 ALK28774.1 DQ243976 ABB90519.1 DQ243979 ABB90522.1 KF514432 AGT21360.1 DQ243973 DQ243974 DQ243975 ABB90516.1 ABB90517.1 ABB90518.1 KF514433 AGT21367.1
MG916903 QBP43279.1 KJ473810 AIA62271.1 JQ065046 AFD29217.1 KY073747 APD51499.1 EF434381 ABO88151.1 KJ473807 AIA62252.1 LC364344 BBC54842.1 LC364343 BBC54832.1 KY073744 APD51475.1 JQ065045 AFD29209.1 HQ728485 ADX59488.1 KY073748 APD51507.1 LC364342 BBC54822.1 KT253267 ALK28773.1 KT253265 ALK28771.1 KT253261 KT253263 ALK28767.1 ALK28769.1 KT253262 ALK28768.1 MK334044 QED88019.1 KT381875 KU521535 ALJ53431.1 ANU06084.1 JX504050 AFV53148.1 AY567487 AY518894 AY758297 AY758298 AY758300 AY758301 KM055634 AIW52831.1 KM055642 AIW52839.1 MK334043 QED88012.1 MN306040 QEG03748.1 JX104161 JX524171 KM055632 KM055633 KM055635 KM055636 KM055637 KM055648 KM055649 KM055650 MG428699 MG428700 MG428701 MG428702 MG428703 MG428704 MG428705 MG428706 AFO70497.1 AFQ55884.1 AIW52829.1 AIW52830.1 AIW52832.1 AIW52833.1 AIW52834.1 AIW52845.1 AIW52846.1 AIW52847.1 AWK59931.1 AWK59937.1 AWK59943.1 AWK59949.1 AWK59955.1 AWK59961.1 AWK59967.1 AWK59973.1 KM055644 AIW52841.1 KM055639 KM055640 KM055641 KM055645 KM055647 AIW52836.1 AIW52837.1 AIW52838.1 AIW52842.1 AIW52844.1 KY554970 ARE29979.1 KY674916 ARB07406.1 JQ765567 AFD98778.1 KT266906 KX179494 KX179495 KX179496 AKT07952.1 APF29061.1 APF29062.1 APF29063.1 KY674915 KY554967 ARB07399.1 ARE29961.1 JQ771055 JQ771057 JQ771058 AFD64748.1 AFD64754.1 AFD64757.1 KT253270 ALK28781.1 KF530107 AGT51345.1 DQ445911 ABE97137.1 KM055638 AIW52835.1 MK334045 QED88026.1 KX179500 APF29071.1 MG428707 AWK59979.1 JQ765574 AFD98827.1 KM055643 AIW52840.1 KF530104 KF530106 KF530108 KF530109 KF530111 KF530113 KF530114 AGT51324.1 AGT51338.1 AGT51352.1 AGT51359.1 AGT51373.1 AGT51387.1 AGT51394.1 JQ765575 AFD98834.1 JQ765568 AFD98785.1 KF530105 AGT51331.1 JQ765571 JQ765572 AFD98806.1 AFD98813.1 JQ771059 AFD64760.1 KY554968 ARE29967.1 JQ765573 AFD98820.1 KY554969 KY554971 KY829118 ARE29973.1 ARE29985.1 ARK08627.1 KT368916 ALA50256.1 KM055646 AIW52843.1 KF530112 AGT51380.1 MN026166 QDH43736.1 MF996621 MF996622 AUF73167.1 KF530110 AGT51366.1 JQ765566 AFD98771.1 MG772808 AVA26873.1 KT368915 ALA50249.1 KT368899 ALA50137.1 KT368901 ALA50151.1 KT368907 KT368908 ALA50193.1 ALA50200.1 KT253327 AOI28283.1 KT368898 ALA50130.1 KT368903 ALA50165.1 KU291449 AOG74789.1 KT368892 KT368893 KT368894 KT368902 KT368904 KT368905 KT368906 KT368909 KT368910 KT368911 KT368912 KT368913 KT368914 ALA50088.1 ALA50095.1 ALA50102.1 ALA50158.1 ALA50172.1 ALA50179.1 ALA50186.1 ALA50207.1 ALA50214.1 ALA50221.1 ALA50228.1 ALA50235.1 ALA50242.1 MK720945 MK720946 QCX35167.1 QCX35178.1 KT253324 KT253326 KT253328 AOI28259.1 AOI28275.1 AOI28291.1 MK334046 QED88033.1 JQ410000 AFI49431.1 JQ765563 AFD98750.1 KY983586 ARU07594.1 KT253325 AOI28267.1 DQ445912 JQ771056 JQ771060 JQ765564 JQ765565 JQ765569 JQ765570 ABE97130.1 AFD64751.1 AFD64763.1 AFD98757.1 AFD98764.1 AFD98792.1 AFD98799.1 MF996623 AUF73168.1 KT368895 KT368896 MF593473 ALA50109.1 ALA50116.1 ATI09437.1 X16816 AF304460 AF344186 AF344187 AF344188 AF344189 Y09923 Y10051 Y10052 X15654 KT368900 ALA50144.1 KT368897 ALA50123.1 KT253266 ALK28772.1 KF293664 KF293665 KF293666 AGW80948.1 DQ243963 ABB90506.1 AB691763 BAL45637.1 KT253269 ALK28775.1 MN306018 QEG03731.1 KT253271 ALK28787.1 KT253264 ALK28770.1 MK334047 QED88040.1 KT253268 ALK28774.1 DQ243976 ABB90519.1 DQ243979 ABB90522.1 KF514432 AGT21360.1 DQ243973 DQ243974 DQ243975 ABB90516.1 ABB90517.1 ABB90518.1 KF514433 AGT21367.1
Proteomes
UP000167590
UP000144177
UP000162232
UP000106394
UP000204102
UP000142997
+ More
UP000145724 UP000103541 UP000008573 UP000161757 UP000098690 UP000158277 UP000125140 UP000141634 UP000122373 UP000174240 UP000115484 UP000129674 UP000106901 UP000140530 UP000144638 UP000146557 UP000153644 UP000162862 UP000167796 UP000110792 UP000168590 UP000171901 UP000134798 UP000141321 UP000100468 UP000152663 UP000136929 UP000243277 UP000143818 UP000108941 UP000153625 UP000136947 UP000173458 UP000120649 UP000151210 UP000116691 UP000103783 UP000100137 UP000103248 UP000108758 UP000110675 UP000115199 UP000115399 UP000124352 UP000128900 UP000136940 UP000152773 UP000153580 UP000171347 UP000173795 UP000146121 UP000173136 UP000105969 UP000130385 UP000138462 UP000151180 UP000151840 UP000101679 UP000169049 UP000006716 UP000104585 UP000147133 UP000118770 UP000096398 UP000110267 UP000139984
UP000145724 UP000103541 UP000008573 UP000161757 UP000098690 UP000158277 UP000125140 UP000141634 UP000122373 UP000174240 UP000115484 UP000129674 UP000106901 UP000140530 UP000144638 UP000146557 UP000153644 UP000162862 UP000167796 UP000110792 UP000168590 UP000171901 UP000134798 UP000141321 UP000100468 UP000152663 UP000136929 UP000243277 UP000143818 UP000108941 UP000153625 UP000136947 UP000173458 UP000120649 UP000151210 UP000116691 UP000103783 UP000100137 UP000103248 UP000108758 UP000110675 UP000115199 UP000115399 UP000124352 UP000128900 UP000136940 UP000152773 UP000153580 UP000171347 UP000173795 UP000146121 UP000173136 UP000105969 UP000130385 UP000138462 UP000151180 UP000151840 UP000101679 UP000169049 UP000006716 UP000104585 UP000147133 UP000118770 UP000096398 UP000110267 UP000139984
PRIDE
ProteinModelPortal
H9BR25
A0A2U9I6C2
A0A482JX42
A0A482JZV2
A0A0U1UZ37
H9BR08
+ More
A0A1L2KGE4 A4ULL1 A0A0U1WJW2 A0A2Z5XXT6 A0A2Z5XXS4 A0A1L2KGB2 H9BR00 F1DB14 A0A1L2KGF1 A0A2Z5XXR5 A0A0P0KBC5 A0A0P0K0X5 A0A0N7J128 A0A0P0KXK6 A0A5B9BH95 A0A0P0G321 K4P2Y7 Q6Q1S2 A0A384RHS8 A0A384QZE9 A0A5B9BIJ4 A0A5B9MUE4 I7CMD1 A0A384RHT2 A0A384QXZ7 A0A1V0PKX8 A0A1V0E0U5 H9EJ44 A0A0K1D6F6 A0A1V0E0W0 H9EJV2 A0A0P0KR98 U3M7G5 Q06Y16 A0A384RBV6 A0A5B9BH24 A0A1L2YVI8 A0A2U8JDM5 H9EJ93 A0A384RK30 U3M6U5 H9EJA0 H9EJ51 U3M6Q4 H9EJ72 H9EJW4 A0A1V0PKW6 H9EJ86 A0A1V0PKX1 A0A0U2GNG3 A0A384R2Q9 U3M7H2 A0A513ZT30 A0A2H4ZBM8 U3M6S0 H9EJ37 A0A2L0WQE1 A0A0U2GQI5 A0A0U2GQ76 A0A0U2GMQ4 A0A0U2GRB2 A0A1C8YZ34 A0A0U2GN50 A0A0U2GRL6 A0A1B3TNA4 A0A0U2GMN8 A0A4Y5QL66 A0A1C8YZ37 A0A5B9BHU7 I1VWF5 H9EJ16 A0A1Y0EV12 A0A1C8YZ26 Q06Y10 A0A2H4ZBN0 A0A0U2GME6 P15423 A0A0U2GN82 A0A0U2GMG2 A0A0P0KI80 A0A059SGA9 Q1HVM6 H1AG29 A0A0N7J129 A0A5B9MSJ9 A0A0P0K6L9 A0A0P0KR90 A0A5B9BGI8 A0A0P0KMM1 Q1HVL3 Q1HVL0 S5YGU2 Q1HVL4 S5YAI3
A0A1L2KGE4 A4ULL1 A0A0U1WJW2 A0A2Z5XXT6 A0A2Z5XXS4 A0A1L2KGB2 H9BR00 F1DB14 A0A1L2KGF1 A0A2Z5XXR5 A0A0P0KBC5 A0A0P0K0X5 A0A0N7J128 A0A0P0KXK6 A0A5B9BH95 A0A0P0G321 K4P2Y7 Q6Q1S2 A0A384RHS8 A0A384QZE9 A0A5B9BIJ4 A0A5B9MUE4 I7CMD1 A0A384RHT2 A0A384QXZ7 A0A1V0PKX8 A0A1V0E0U5 H9EJ44 A0A0K1D6F6 A0A1V0E0W0 H9EJV2 A0A0P0KR98 U3M7G5 Q06Y16 A0A384RBV6 A0A5B9BH24 A0A1L2YVI8 A0A2U8JDM5 H9EJ93 A0A384RK30 U3M6U5 H9EJA0 H9EJ51 U3M6Q4 H9EJ72 H9EJW4 A0A1V0PKW6 H9EJ86 A0A1V0PKX1 A0A0U2GNG3 A0A384R2Q9 U3M7H2 A0A513ZT30 A0A2H4ZBM8 U3M6S0 H9EJ37 A0A2L0WQE1 A0A0U2GQI5 A0A0U2GQ76 A0A0U2GMQ4 A0A0U2GRB2 A0A1C8YZ34 A0A0U2GN50 A0A0U2GRL6 A0A1B3TNA4 A0A0U2GMN8 A0A4Y5QL66 A0A1C8YZ37 A0A5B9BHU7 I1VWF5 H9EJ16 A0A1Y0EV12 A0A1C8YZ26 Q06Y10 A0A2H4ZBN0 A0A0U2GME6 P15423 A0A0U2GN82 A0A0U2GMG2 A0A0P0KI80 A0A059SGA9 Q1HVM6 H1AG29 A0A0N7J129 A0A5B9MSJ9 A0A0P0K6L9 A0A0P0KR90 A0A5B9BGI8 A0A0P0KMM1 Q1HVL3 Q1HVL0 S5YGU2 Q1HVL4 S5YAI3
PDB
5SZS
E-value=0,
Score=2217
Ontologies
GO
GO:0016021 C:integral component of membrane
GO:0019031 C:viral envelope
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0046789 F:host cell surface receptor binding
GO:0019031 C:viral envelope
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0046789 F:host cell surface receptor binding
Subcellular Location
From MSLVP
Single-Pass Membrane
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
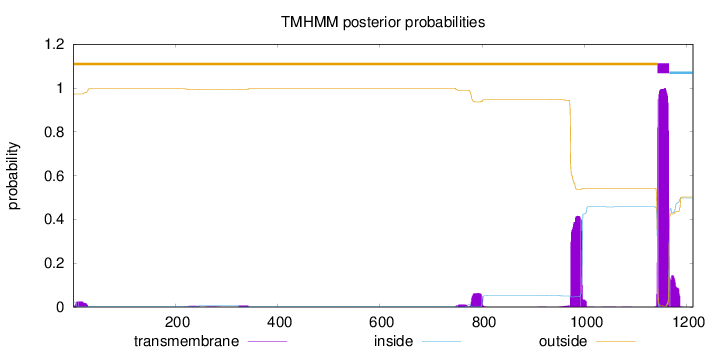
Topology
Length:
1212
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
37.09279
Exp number, first 60 AAs:
0.52552
Total prob of N-in:
0.02752
outside
1 - 1142
TMhelix
1143 - 1165
inside
1166 - 1212
Population Genetic Test Statistics
Pi
0.4628237
Theta
0.5876789
Tajima's D
-0.8644114
CLR
7.218802
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 |
Protein |
YP_005352871.1 |
|
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.