Strain
USA_MURINE_1998_NC_023760
(Region: USA: Wisconsin; Strain: Mink coronavirus strain WD1127, complete genome.; Date: 1998)
Gene
envelope protein
Description
Annotated in NCBI,
envelope protein
Location
GenBank Accession
Full name
Envelope small membrane protein
Sequence
CDS
ATGAGATTCCCGAACGTCTTGACTGTGATTGATGACAATGGAATTGTAATCAACTCCATATTTTGGCTACTGTTGATAATTGTAATTATATTATTTTCTATAGCACTACTTAACGTTGTTAAGTTGTGCATGGTTTGTTGTAGATTAAGCAATGTTGTTATCATTGCGCCTGCTCGACAAGCCTATAATGCCTATAAAGATTTCATGAACATACCAAAAGCACCAGACAGTGTTTGTATTGTTGTTTAA
Protein
MRFPNVLTVIDDNGIVINSIFWLLLIIVIILFSIALLNVVKLCMVCCRLSNVVIIAPARQAYNAYKDFMNIPKAPDSVCIVV
Summary
Function
Plays a central role in virus morphogenesis and assembly. Acts as a viroporin and self-assembles in host membranes forming pentameric protein-lipid pores that allow ion transport. Also plays a role in the induction of apoptosis.
Subunit
Homopentamer. Interacts with membrane protein M in the budding compartment of the host cell, which is located between endoplasmic reticulum and the Golgi complex. Interacts with Nucleoprotein.
Similarity
Belongs to the alphacoronaviruses E protein family.
Keywords
Apoptosis
Host Golgi apparatus
Host membrane
Membrane
Transmembrane
Transmembrane helix
Reference proteome
Feature
chain Envelope small membrane protein
Uniprot
D9J1Z6
A0A2R4QL30
D9J205
A0A222UCB7
A0A1B4X969
D3XGJ6
+ More
A0A172B5K4 A0A224ANV7 A0A222UC56 D3XGI8 A0A0K2SFA3 Q6Q2E0 P36696 S6BPG0 S6BUM6 I0B0R4 G8J7G8 A0A088ND87 C6GHG0 W0U135 A0A386YL30 W0U130 A0A088NBY1 A0A088NCW5 W0U1A4 Q6Q2D9 C6GHJ3 C6GHI2 A0A125R5H5 A0A2U8Z8V4 C5IJ75 A0A515KYV6 A0A088NFR3 A3DT39 O39435 A0A2H5BX06 C6GH88 E0AGZ0 E0AGV9 A0A088NBY7 T1SR97 E3W5G1 E3W5J3 A0A088NS11 C5IGF2 E0AH01 E3W5I3 A0A088NFY5 C6GHC9 D3KDL8 C6GHB0 E3W5H2 D3KDK7 Q3Y4E7 C6GHH1 A0A2H4KI33 C6GHD9 G1FEZ0 A0A1Z2WR54 A1DS44 A0A088NBU4 A0A088NC56 G1FEX9 C6GHB9 E0AGX9 A0A077JGD3 K7YHT2 V9PIT6 Q52PA5 B7T075 C5IG90 A0A088NRV6 D6RTP3 E0AGU8 C5IJ37 C6GHK4 C5IGA8 I0B0S3 C6K2T9 O12296 Q0ZVX2 E0AGW9 C5IGE3 K7Y4N5 A0A3Q8BZD0 A0A088NBX6
A0A172B5K4 A0A224ANV7 A0A222UC56 D3XGI8 A0A0K2SFA3 Q6Q2E0 P36696 S6BPG0 S6BUM6 I0B0R4 G8J7G8 A0A088ND87 C6GHG0 W0U135 A0A386YL30 W0U130 A0A088NBY1 A0A088NCW5 W0U1A4 Q6Q2D9 C6GHJ3 C6GHI2 A0A125R5H5 A0A2U8Z8V4 C5IJ75 A0A515KYV6 A0A088NFR3 A3DT39 O39435 A0A2H5BX06 C6GH88 E0AGZ0 E0AGV9 A0A088NBY7 T1SR97 E3W5G1 E3W5J3 A0A088NS11 C5IGF2 E0AH01 E3W5I3 A0A088NFY5 C6GHC9 D3KDL8 C6GHB0 E3W5H2 D3KDK7 Q3Y4E7 C6GHH1 A0A2H4KI33 C6GHD9 G1FEZ0 A0A1Z2WR54 A1DS44 A0A088NBU4 A0A088NC56 G1FEX9 C6GHB9 E0AGX9 A0A077JGD3 K7YHT2 V9PIT6 Q52PA5 B7T075 C5IG90 A0A088NRV6 D6RTP3 E0AGU8 C5IJ37 C6GHK4 C5IGA8 I0B0S3 C6K2T9 O12296 Q0ZVX2 E0AGW9 C5IGE3 K7Y4N5 A0A3Q8BZD0 A0A088NBX6
Pubmed
EMBL
HM245925
ADI80515.1
MF113046
AVY53337.1
HM245926
ADI80524.1
+ More
KX512809 ASR18940.1 LC119077 BAV31351.1 GU338457 ADD49360.1 KM347965 AKG92642.1 LC215871 BBA21170.1 KX512810 ASR18948.1 GU338456 ADD49351.1 LC029419 BAS25712.1 AY566160 AAS68509.1 D13096 AB781788 BAN67904.1 AB781790 BAN67922.1 JQ408980 AFH55115.1 JN856008 AEQ61971.1 KJ879390 AIN55906.1 FJ938058 ACT10930.1 HE860490 CCI69283.1 MH817484 AYF53097.1 KP143507 KP143508 KP143509 KP143510 KP143511 KP143512 HE598741 HE598742 HE598743 HE598744 HE598745 HE598746 HE598747 HE860479 HE860480 HE860481 HE860482 HE860483 HE860484 HE860485 HE860487 HE860488 HE860489 HE860491 AJO26977.1 AJO26987.1 AJO26997.1 AJO27007.1 AJO27017.1 AJO27027.1 CCD41999.1 CCI69261.1 CCI69263.1 CCI69265.1 CCI69267.1 CCI69269.1 CCI69271.1 CCI69273.1 CCI69277.1 CCI69279.1 CCI69281.1 CCI69285.1 KJ879347 KJ879387 KJ879388 AIN55788.1 AIN55893.1 AIN55899.1 KJ879340 KJ879341 KJ879342 KJ879352 KJ879391 KJ879392 KJ879393 AIN55771.1 AIN55775.1 AIN55776.1 AIN55797.1 AIN55910.1 AIN55913.1 AIN55914.1 HE860486 CCI69275.1 AY566161 JQ404409 KC175340 AAS68510.1 AFG19731.1 AFX81101.1 FJ938061 ACT10963.1 FJ938060 ACT10952.1 KU215419 KU215420 KU215421 KU215422 KU215423 KU215424 KU215425 KU215426 KU215427 KU215428 AMD11134.1 AMD11146.1 AMD11157.1 AMD11168.1 AMD11179.1 AMD11190.1 AMD11201.1 AMD11212.1 AMD11223.1 AMD11234.1 MF457591 AWN93245.1 FJ943771 ACR46151.1 MN165107 QDM36990.1 KJ879375 KJ879376 KJ879378 AIN55844.1 AIN55849.1 AIN55857.1 DQ286389 ABD33843.1 AF033000 KC461235 AAB87471.1 AGZ84520.1 KY566209 KY566210 KY566211 AUG98123.1 AUG98134.1 AUG98145.1 FJ938051 ACT10858.1 HQ012371 ADL71490.1 HQ012368 ADL71459.1 KJ879348 KJ879349 KJ879350 KJ879385 AIN55789.1 AIN55792.1 AIN55793.1 AIN55885.1 KF530123 AGT52084.1 HQ392469 ADO39821.1 HQ392472 ADO39853.1 KJ879386 AIN55889.1 FJ917526 FJ943762 ACR10345.1 ACR46121.1 HQ012372 ADL71501.1 HQ392471 ADO39843.1 KJ879389 AIN55902.1 FJ938055 ACT10899.1 GU553362 ADC35476.1 FJ938053 ACT10880.1 HQ392470 ADO39832.1 GU553361 ADC35465.1 DQ160294 EU186072 KJ879335 KJ879336 KJ879337 KJ879338 KJ879339 KJ879351 KY292377 KX722529 AAZ86081.1 ABX60146.1 AIN55757.1 AIN55761.1 AIN55762.1 AIN55767.1 AIN55770.1 AIN55795.1 ASB16891.1 ASU62492.1 FJ938059 ACT10941.1 KX722530 KX722531 ASU62503.1 ASU62513.1 FJ938056 ACT10909.1 JN183883 AEK25525.1 KY063616 KY063617 KY063618 ASB15742.1 ASB15752.1 ASB15760.1 EF056487 ABK79900.1 KJ879334 AIN55753.1 KJ879377 AIN55853.1 JN183882 AEK25514.1 FJ938054 ACT10889.1 HQ012370 ADL71479.1 AB907624 BAP19070.1 KC175339 AFX81094.1 KC461236 KC461237 AGZ84530.1 AGZ84540.1 DQ010921 AY994055 EU856362 ACJ63246.1 FJ917519 FJ917520 FJ917530 FJ917531 FJ917532 FJ917533 FJ917534 FJ917535 ACR10283.1 ACR10292.1 ACR10381.1 ACR10390.1 ACR10399.1 ACR10408.1 ACR10417.1 ACR10425.1 KJ879343 KJ879344 KJ879346 KJ879379 KJ879380 KJ879381 KJ879382 KJ879383 KJ879384 AIN55779.1 AIN55781.1 AIN55785.1 AIN55860.1 AIN55865.1 AIN55869.1 AIN55873.1 AIN55876.1 AIN55881.1 AB535528 BAJ08259.1 HQ012367 ADL71448.1 FJ943761 ACR46113.1 FJ938062 ACT10974.1 FJ917521 FJ917522 FJ917524 FJ943763 ACR10301.1 ACR10310.1 ACR10328.1 ACR46124.1 JQ408981 AFH55124.1 GQ152141 ACS44221.1 Y13921 CAA74228.1 DQ112226 EU856361 EU924791 GU146061 GQ477367 HQ450376 JF682842 KP981644 AAZ91440.1 ACJ63235.1 ACJ64188.1 ADB10853.1 ADB28911.1 ADU17729.1 AEE69326.1 AKA65833.1 HQ012369 ADL71469.1 FJ917525 ACR10336.1 KC175341 AFX81111.1 KY406735 ASV45151.1 KJ879345 AIN55783.1
KX512809 ASR18940.1 LC119077 BAV31351.1 GU338457 ADD49360.1 KM347965 AKG92642.1 LC215871 BBA21170.1 KX512810 ASR18948.1 GU338456 ADD49351.1 LC029419 BAS25712.1 AY566160 AAS68509.1 D13096 AB781788 BAN67904.1 AB781790 BAN67922.1 JQ408980 AFH55115.1 JN856008 AEQ61971.1 KJ879390 AIN55906.1 FJ938058 ACT10930.1 HE860490 CCI69283.1 MH817484 AYF53097.1 KP143507 KP143508 KP143509 KP143510 KP143511 KP143512 HE598741 HE598742 HE598743 HE598744 HE598745 HE598746 HE598747 HE860479 HE860480 HE860481 HE860482 HE860483 HE860484 HE860485 HE860487 HE860488 HE860489 HE860491 AJO26977.1 AJO26987.1 AJO26997.1 AJO27007.1 AJO27017.1 AJO27027.1 CCD41999.1 CCI69261.1 CCI69263.1 CCI69265.1 CCI69267.1 CCI69269.1 CCI69271.1 CCI69273.1 CCI69277.1 CCI69279.1 CCI69281.1 CCI69285.1 KJ879347 KJ879387 KJ879388 AIN55788.1 AIN55893.1 AIN55899.1 KJ879340 KJ879341 KJ879342 KJ879352 KJ879391 KJ879392 KJ879393 AIN55771.1 AIN55775.1 AIN55776.1 AIN55797.1 AIN55910.1 AIN55913.1 AIN55914.1 HE860486 CCI69275.1 AY566161 JQ404409 KC175340 AAS68510.1 AFG19731.1 AFX81101.1 FJ938061 ACT10963.1 FJ938060 ACT10952.1 KU215419 KU215420 KU215421 KU215422 KU215423 KU215424 KU215425 KU215426 KU215427 KU215428 AMD11134.1 AMD11146.1 AMD11157.1 AMD11168.1 AMD11179.1 AMD11190.1 AMD11201.1 AMD11212.1 AMD11223.1 AMD11234.1 MF457591 AWN93245.1 FJ943771 ACR46151.1 MN165107 QDM36990.1 KJ879375 KJ879376 KJ879378 AIN55844.1 AIN55849.1 AIN55857.1 DQ286389 ABD33843.1 AF033000 KC461235 AAB87471.1 AGZ84520.1 KY566209 KY566210 KY566211 AUG98123.1 AUG98134.1 AUG98145.1 FJ938051 ACT10858.1 HQ012371 ADL71490.1 HQ012368 ADL71459.1 KJ879348 KJ879349 KJ879350 KJ879385 AIN55789.1 AIN55792.1 AIN55793.1 AIN55885.1 KF530123 AGT52084.1 HQ392469 ADO39821.1 HQ392472 ADO39853.1 KJ879386 AIN55889.1 FJ917526 FJ943762 ACR10345.1 ACR46121.1 HQ012372 ADL71501.1 HQ392471 ADO39843.1 KJ879389 AIN55902.1 FJ938055 ACT10899.1 GU553362 ADC35476.1 FJ938053 ACT10880.1 HQ392470 ADO39832.1 GU553361 ADC35465.1 DQ160294 EU186072 KJ879335 KJ879336 KJ879337 KJ879338 KJ879339 KJ879351 KY292377 KX722529 AAZ86081.1 ABX60146.1 AIN55757.1 AIN55761.1 AIN55762.1 AIN55767.1 AIN55770.1 AIN55795.1 ASB16891.1 ASU62492.1 FJ938059 ACT10941.1 KX722530 KX722531 ASU62503.1 ASU62513.1 FJ938056 ACT10909.1 JN183883 AEK25525.1 KY063616 KY063617 KY063618 ASB15742.1 ASB15752.1 ASB15760.1 EF056487 ABK79900.1 KJ879334 AIN55753.1 KJ879377 AIN55853.1 JN183882 AEK25514.1 FJ938054 ACT10889.1 HQ012370 ADL71479.1 AB907624 BAP19070.1 KC175339 AFX81094.1 KC461236 KC461237 AGZ84530.1 AGZ84540.1 DQ010921 AY994055 EU856362 ACJ63246.1 FJ917519 FJ917520 FJ917530 FJ917531 FJ917532 FJ917533 FJ917534 FJ917535 ACR10283.1 ACR10292.1 ACR10381.1 ACR10390.1 ACR10399.1 ACR10408.1 ACR10417.1 ACR10425.1 KJ879343 KJ879344 KJ879346 KJ879379 KJ879380 KJ879381 KJ879382 KJ879383 KJ879384 AIN55779.1 AIN55781.1 AIN55785.1 AIN55860.1 AIN55865.1 AIN55869.1 AIN55873.1 AIN55876.1 AIN55881.1 AB535528 BAJ08259.1 HQ012367 ADL71448.1 FJ943761 ACR46113.1 FJ938062 ACT10974.1 FJ917521 FJ917522 FJ917524 FJ943763 ACR10301.1 ACR10310.1 ACR10328.1 ACR46124.1 JQ408981 AFH55124.1 GQ152141 ACS44221.1 Y13921 CAA74228.1 DQ112226 EU856361 EU924791 GU146061 GQ477367 HQ450376 JF682842 KP981644 AAZ91440.1 ACJ63235.1 ACJ64188.1 ADB10853.1 ADB28911.1 ADU17729.1 AEE69326.1 AKA65833.1 HQ012369 ADL71469.1 FJ917525 ACR10336.1 KC175341 AFX81111.1 KY406735 ASV45151.1 KJ879345 AIN55783.1
Proteomes
UP000156811
UP000132946
UP000272061
UP000150460
UP000096172
UP000153699
+ More
UP000122558 UP000100168 UP000105943 UP000115718 UP000140032 UP000149467 UP000170923 UP000119169 UP000155481 UP000147290 UP000118118 UP000102801 UP000113596 UP000117806 UP000125896 UP000126919 UP000135072 UP000135717 UP000143834 UP000158395 UP000161862 UP000144800 UP000169693 UP000117062 UP000169984 UP000141579 UP000113324 UP000113038 UP000153548 UP000110620 UP000145457 UP000162682 UP000138585 UP000123646 UP000165438 UP000137357 UP000141821 UP000153160 UP000140550 UP000173564 UP000144443 UP000155736 UP000128709 UP000119918 UP000131699 UP000156095 UP000000835 UP000140386 UP000171330 UP000173643 UP000180339 UP000164750 UP000115330 UP000129066 UP000161789 UP000165378
UP000122558 UP000100168 UP000105943 UP000115718 UP000140032 UP000149467 UP000170923 UP000119169 UP000155481 UP000147290 UP000118118 UP000102801 UP000113596 UP000117806 UP000125896 UP000126919 UP000135072 UP000135717 UP000143834 UP000158395 UP000161862 UP000144800 UP000169693 UP000117062 UP000169984 UP000141579 UP000113324 UP000113038 UP000153548 UP000110620 UP000145457 UP000162682 UP000138585 UP000123646 UP000165438 UP000137357 UP000141821 UP000153160 UP000140550 UP000173564 UP000144443 UP000155736 UP000128709 UP000119918 UP000131699 UP000156095 UP000000835 UP000140386 UP000171330 UP000173643 UP000180339 UP000164750 UP000115330 UP000129066 UP000161789 UP000165378
PRIDE
Pfam
PF02723
NS3_envE
Interpro
IPR003873
NS3/E
ProteinModelPortal
D9J1Z6
A0A2R4QL30
D9J205
A0A222UCB7
A0A1B4X969
D3XGJ6
+ More
A0A172B5K4 A0A224ANV7 A0A222UC56 D3XGI8 A0A0K2SFA3 Q6Q2E0 P36696 S6BPG0 S6BUM6 I0B0R4 G8J7G8 A0A088ND87 C6GHG0 W0U135 A0A386YL30 W0U130 A0A088NBY1 A0A088NCW5 W0U1A4 Q6Q2D9 C6GHJ3 C6GHI2 A0A125R5H5 A0A2U8Z8V4 C5IJ75 A0A515KYV6 A0A088NFR3 A3DT39 O39435 A0A2H5BX06 C6GH88 E0AGZ0 E0AGV9 A0A088NBY7 T1SR97 E3W5G1 E3W5J3 A0A088NS11 C5IGF2 E0AH01 E3W5I3 A0A088NFY5 C6GHC9 D3KDL8 C6GHB0 E3W5H2 D3KDK7 Q3Y4E7 C6GHH1 A0A2H4KI33 C6GHD9 G1FEZ0 A0A1Z2WR54 A1DS44 A0A088NBU4 A0A088NC56 G1FEX9 C6GHB9 E0AGX9 A0A077JGD3 K7YHT2 V9PIT6 Q52PA5 B7T075 C5IG90 A0A088NRV6 D6RTP3 E0AGU8 C5IJ37 C6GHK4 C5IGA8 I0B0S3 C6K2T9 O12296 Q0ZVX2 E0AGW9 C5IGE3 K7Y4N5 A0A3Q8BZD0 A0A088NBX6
A0A172B5K4 A0A224ANV7 A0A222UC56 D3XGI8 A0A0K2SFA3 Q6Q2E0 P36696 S6BPG0 S6BUM6 I0B0R4 G8J7G8 A0A088ND87 C6GHG0 W0U135 A0A386YL30 W0U130 A0A088NBY1 A0A088NCW5 W0U1A4 Q6Q2D9 C6GHJ3 C6GHI2 A0A125R5H5 A0A2U8Z8V4 C5IJ75 A0A515KYV6 A0A088NFR3 A3DT39 O39435 A0A2H5BX06 C6GH88 E0AGZ0 E0AGV9 A0A088NBY7 T1SR97 E3W5G1 E3W5J3 A0A088NS11 C5IGF2 E0AH01 E3W5I3 A0A088NFY5 C6GHC9 D3KDL8 C6GHB0 E3W5H2 D3KDK7 Q3Y4E7 C6GHH1 A0A2H4KI33 C6GHD9 G1FEZ0 A0A1Z2WR54 A1DS44 A0A088NBU4 A0A088NC56 G1FEX9 C6GHB9 E0AGX9 A0A077JGD3 K7YHT2 V9PIT6 Q52PA5 B7T075 C5IG90 A0A088NRV6 D6RTP3 E0AGU8 C5IJ37 C6GHK4 C5IGA8 I0B0S3 C6K2T9 O12296 Q0ZVX2 E0AGW9 C5IGE3 K7Y4N5 A0A3Q8BZD0 A0A088NBX6
Ontologies
GO
Subcellular Location
From MSLVP
Host nucleus. Host cytoplasm.
From Uniprot
Host Golgi apparatus membrane
The cytoplasmic tail functions as a Golgi complex-targeting signal. With evidence from 1 publications.
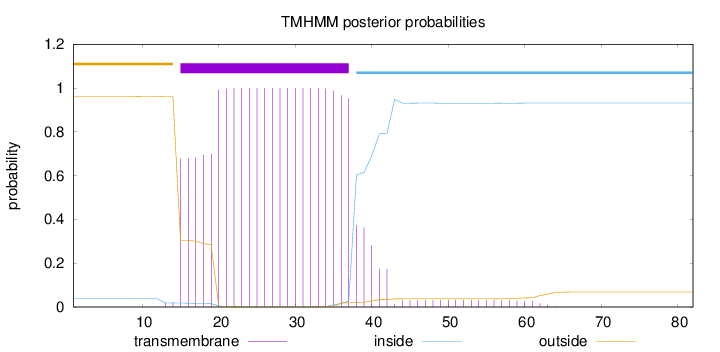
Topology
Length:
82
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.34608
Exp number, first 60 AAs:
23.28789
Total prob of N-in:
0.03775
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 82
Population Genetic Test Statistics
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 |
Protein |
YP_009019184.1 |
|
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 |
Protein |
YP_009256199.1 |
|
| JAPAN_FERRET_2013_LC119077 |
Protein |
BAV31351.1 |
|
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
