Strain
CHINA_MURINE_2012_NC_026011
(Region: China; Strain: Betacoronavirus HKU24 strain HKU24-R05005I, complete genome.; Date: 2012)
Gene
hemagglutinin-esterase protein
Description
Annotated in NCBI,
hemagglutinin-esterase protein
Location
GenBank Accession
Full name
Hemagglutinin-esterase
Alternative Name
E3 glycoprotein
Sequence
CDS
ATGGCCACCGCAGTCCTAACCATAGGCCTGCTTTGCCTCCTCGTCGGTGTTACAGGTTTTGAAAATCCTCCCACCAACGTGGTTTCTCACATTAATGACGATTGGTTTTTGTTTGGCGACAGTCGTAGTGACTGTAACCATATCAATAATATCAATCCTAAAAACTATTCTTATATGGACCTCGATCCCTCGTTGTGCAATTCAGGAAAAATATCTTCTAAGGCTGGCAATTCCATCTTTAGGAGCTTTCATTTCACTGACTTCTACAACTATCAGGGTGAAGGGAACCAAATAATATTCTATGAAGGCGTTAATTTTTCACCGAGTCATGCGTTTAAATGTACCTCGTCCGGTGATAATGGTATTTGGATAAATAATAAAGGTAAGTTTTATGAGCAAGTGTATAAGAAGATGTCCGTTTATCGAGGCCTCTCCATAATAACTGTTCCCTATGTGTACACTGGAGCGGCAGCTGCAACCTCAATTTGTAAGAGGGGCAGTCTGACCTTAAATAATCCAGCGTCTATACAGAGGGAAACTGGTGTTAATACTTATTTTTACACCGCTGAGGCTAATTTTACTCTCACGGGTTGTGATGAGTACATAGTCCCATTGTGTATTTATACTGGTATGTTCCTGGCTTCAGGTACTTACTACAATGACAGCCAGTATTATTATAGTTATAGTACTGGAGAATTCTATGGCCGTAACAACACAGTTGATAAAGCTACTGTAGTTGATTACGAGTGCCAATATCTTACCTTGCCTTCAGGAATTTATAAATCTGTTTCTAATGAGTTGCTTCTCACAGTTCCGACGAAGGCTATCTGTCTGACAAAGCCGAAGAGCTTTACGCCAGTACAGGTTGTCGATTCTCGTTGGAACCATGCCAGGACCTCTGATAATAAAACAGCAGAGGCATGTCAATTGCCTTATTGCTATTTTCGTAATTCTACCACAGATTATGTTGGTAGCTATGATATTAACCATGGCGATGTTGGATTTTCTAACATCCTCAAGGGTTTAATTTATAATGCCCCTTGCTTTGCTAAGCAAGGTGTGTTTATGTATAATAATGTTAGTAGTGTTTGGCCCGTCTATGCCTATGGCGAGTGCCCCACTGCTGCCAATATAAATATAAATGGAATTCCCATTTGTAATTATGATCCGCTACCTGTCATTTTGCTTGGCATTCTTCTTGGAATAGCGGTTATAACCATTGTGGTTTTGCTTTTATATTTCCTTGTAGATAATGGTACTAGGCTTCATGATGCGTAG
Protein
MATAVLTIGLLCLLVGVTGFENPPTNVVSHINDDWFLFGDSRSDCNHINNINPKNYSYMDLDPSLCNSGKISSKAGNSIFRSFHFTDFYNYQGEGNQIIFYEGVNFSPSHAFKCTSSGDNGIWINNKGKFYEQVYKKMSVYRGLSIITVPYVYTGAAAATSICKRGSLTLNNPASIQRETGVNTYFYTAEANFTLTGCDEYIVPLCIYTGMFLASGTYYNDSQYYYSYSTGEFYGRNNTVDKATVVDYECQYLTLPSGIYKSVSNELLLTVPTKAICLTKPKSFTPVQVVDSRWNHARTSDNKTAEACQLPYCYFRNSTTDYVGSYDINHGDVGFSNILKGLIYNAPCFAKQGVFMYNNVSSVWPVYAYGECPTAANININGIPICNYDPLPVILLGILLGIAVITIVVLLLYFLVDNGTRLHDA
Summary
Function
Structural protein that makes short spikes at the surface of the virus. Contains receptor binding and receptor-destroying activities. Mediates de-O-acetylation of N-acetyl-4-O-acetylneuraminic acid, which is probably the receptor determinant recognized by the virus on the surface of erythrocytes and susceptible cells. This receptor-destroying activity is important for virus release as it probably helps preventing self-aggregation and ensures the efficient spread of the progeny virus from cell to cell. May serve as a secondary viral attachment protein for initiating infection, the spike protein being the major one. May become a target for both the humoral and the cellular branches of the immune system.
Catalytic Activity
H2O + N-acetyl-4-O-acetylneuraminate = acetate + H(+) + N-acetylneuraminate
H2O + N-acetyl-9-O-acetylneuraminate = acetate + H(+) + N-acetylneuraminate
H2O + N-acetyl-9-O-acetylneuraminate = acetate + H(+) + N-acetylneuraminate
Subunit
Homodimer; disulfide-linked. Forms a complex with the M protein in the pre-Golgi. Associates then with S-M complex to form a ternary complex S-M-HE.
Miscellaneous
The sequence shown is that of Quebec reference strain.
Similarity
Belongs to the influenza type C/coronaviruses hemagglutinin-esterase family.
Keywords
3D-structure
Direct protein sequencing
Disulfide bond
Glycoprotein
Hemagglutinin
Host cell membrane
Host membrane
Hydrolase
Membrane
Signal
Transmembrane
Transmembrane helix
Viral envelope protein
Virion
Feature
chain Hemagglutinin-esterase
Uniprot
A0A0A7UX90
A0A0A7V3Y4
A0A0A7V3Z7
A0A3G3NHC2
A0A3G3NH50
A0A3G3NHN3
+ More
A0A2H4MXC4 A0A096XNF9 A0A3G3NHC5 A0A2H4MX16 A0A2H4MZ34 A0A5J6NRP2 A0A191URZ4 A0A5J6NUD6 A0A481SAP1 A0A5J6NRK9 A0A5J6NRQ9 C6GHQ9 B7TYG8 P15776 Q66165 Q2QKN4 A0A451ETZ3 D0EY82 B9TXU2 D0EY92 P59709 P59710 C6GHL2 Q0PL40 A0A451ETZ0 A0A493R5L6 A0A3S9Y2C0 B7U2R4 B7U2N2 B7U2M1 H9AA54 H9AA33 B7U2L0 B7U2S6 D0EY72 A0A3S9Y2S0 A0A2R4STV0 A7BKC1 A0A3Q9K3J3 A0A346I7H2 P31613 A0A1S6KXR2 A0A5J6NRI4 B7U2Q3 H9AA43 A0A3S9Y2A6 A0A3S7GXV6 Q2EEZ7 A0A0U2GN17 A0A291L0R5 B8RIQ6 A0A411D561 A0A3S7GY11 Q9QAQ9 A0A191URB1 C6GHM4 C6GHN6 H9AA64 A4ZU29 A0A1V0JAY6 A6YD39 X2JFB2 A8HB15 A0A2P1IQB6 A4ZU73 A4ZU62 B7U2P3 A4ZU40 A4ZU51 A0A2R4STW8 A0A1J3 Q2EEZ0 Q9DR81 P59711 A0A346I7H1 A4ZTX4 Q2EEZ3 A0A481S1W3 X2JHM7 A4ZU07 Q4U3S2 A4ZTZ6 A0A346I7H4 A4ZU18 A3E2F1 Q2EEZ4 A0A481S1A3 Q4U3S3 A4ZTY5 Q2EEZ2 A0A481S2D5 A0A1J2 Q8V437 Q06BD8
A0A2H4MXC4 A0A096XNF9 A0A3G3NHC5 A0A2H4MX16 A0A2H4MZ34 A0A5J6NRP2 A0A191URZ4 A0A5J6NUD6 A0A481SAP1 A0A5J6NRK9 A0A5J6NRQ9 C6GHQ9 B7TYG8 P15776 Q66165 Q2QKN4 A0A451ETZ3 D0EY82 B9TXU2 D0EY92 P59709 P59710 C6GHL2 Q0PL40 A0A451ETZ0 A0A493R5L6 A0A3S9Y2C0 B7U2R4 B7U2N2 B7U2M1 H9AA54 H9AA33 B7U2L0 B7U2S6 D0EY72 A0A3S9Y2S0 A0A2R4STV0 A7BKC1 A0A3Q9K3J3 A0A346I7H2 P31613 A0A1S6KXR2 A0A5J6NRI4 B7U2Q3 H9AA43 A0A3S9Y2A6 A0A3S7GXV6 Q2EEZ7 A0A0U2GN17 A0A291L0R5 B8RIQ6 A0A411D561 A0A3S7GY11 Q9QAQ9 A0A191URB1 C6GHM4 C6GHN6 H9AA64 A4ZU29 A0A1V0JAY6 A6YD39 X2JFB2 A8HB15 A0A2P1IQB6 A4ZU73 A4ZU62 B7U2P3 A4ZU40 A4ZU51 A0A2R4STW8 A0A1J3 Q2EEZ0 Q9DR81 P59711 A0A346I7H1 A4ZTX4 Q2EEZ3 A0A481S1W3 X2JHM7 A4ZU07 Q4U3S2 A4ZTZ6 A0A346I7H4 A4ZU18 A3E2F1 Q2EEZ4 A0A481S1A3 Q4U3S3 A4ZTY5 Q2EEZ2 A0A481S2D5 A0A1J2 Q8V437 Q06BD8
EC Number
3.1.1.53
Pubmed
25552712
30285857
25463600
31534035
2319653
2103108
+ More
2732694 15507445 18550812 1727608 16809333 23405345 2732684 7730812 11774537 11504427 11376845 11774548 1962455 30771080 18842722 22398294 17374765 9778786 17803510 26678874 19162098 27274850 17434558 17669602 24655427 17916374 29508550 17344285 16954245 20880639 11714968 16604443 17092595 17459444
2732694 15507445 18550812 1727608 16809333 23405345 2732684 7730812 11774537 11504427 11376845 11774548 1962455 30771080 18842722 22398294 17374765 9778786 17803510 26678874 19162098 27274850 17434558 17669602 24655427 17916374 29508550 17344285 16954245 20880639 11714968 16604443 17092595 17459444
EMBL
KM349742
AJA91195.1
KM349743
AJA91206.1
KM349744
AJA91216.1
+ More
MH687969 AYR18606.1 MH687973 MH687974 MH687977 MH687978 AYR18642.1 AYR18651.1 AYR18678.1 AYR18687.1 MH687971 MH687972 AYR18624.1 AYR18633.1 KY370047 ATP66749.1 KF294357 AID16630.1 MH687975 AYR18660.1 KY370052 ATP66778.1 KY370046 ATP66743.1 MN514966 QEY10656.1 KU886219 ANJ04973.1 MN514963 QEY10632.1 MH475914 QBH22432.1 MN514964 MN514965 QEY10640.1 QEY10648.1 MN514967 QEY10664.1 FJ938067 ACT11029.1 FJ415324 ACJ35485.1 U00735 S50936 M80842 DQ011855 AAY68296.1 MK095145 MK095146 AZS64192.1 AZS64193.1 GQ918142 JX860640 ACX46849.1 AFW97359.1 EU401979 ACB30199.1 GQ918143 ACX46859.1 L38962 L38963 U06093 AF239306 AF239307 AF230523 AF230524 AF230525 AF230526 AF230527 AF230528 AF339836 AF220295 S69264 M76372 FJ938063 ACT10982.1 DQ811784 ABG89283.1 MK095135 MK095136 MK095137 AZS64182.1 AZS64183.1 AZS64184.1 MK045995 QBG67064.1 MK095140 AZS64187.1 FJ425189 ACJ66999.1 FJ425186 ACJ66968.1 FJ425185 ACJ66956.1 JN874561 AFE48816.1 JN874559 AFE48794.1 FJ425184 ACJ66944.1 FJ425190 ACJ67011.1 GQ918141 ACX46839.1 MK095147 AZS64194.1 MH043952 AVZ61098.1 EU401977 EU401978 AB354579 ACB30197.1 ACB30198.1 BAF75630.1 MK095138 MK095139 AZS64185.1 AZS64186.1 MH203060 MH203061 MH203063 AXP11692.1 AXP11693.1 AXP11695.1 M84486 AF058942 KX432213 AQT26497.1 MN514962 QEY10624.1 FJ425188 ACJ66986.1 JN874560 AFE48804.1 MK095142 AZS64189.1 MG757138 AVI14999.1 DQ389643 DQ389644 DQ389645 ABD18919.1 ABD18920.1 ABD18921.1 KT368891 ALA50079.1 MF593476 ATI09448.1 EU999954 ACL12995.1 MH249786 QAY30019.1 MG757139 MG757141 AVI15010.1 AVI15032.1 AF058944 KU558922 ANJ04716.1 FJ938064 ACT10994.1 FJ938065 ACT11006.1 JN874562 AFE48826.1 EF424620 ABP38286.1 KX982264 ARD05998.1 EF445634 ABP73655.1 KF906250 KF906251 AHN64782.1 AHN64791.1 EU019216 ABV74053.1 MG518518 AVN88333.1 EF424624 ABP38338.1 EF424623 ABP38323.1 FJ425187 ACJ66975.1 EF424621 ABP38304.1 EF424622 ABP38312.1 MH043955 AVZ61128.1 DQ994168 ABI98029.1 DQ389651 EU401975 EU401976 HM573321 HM573322 HM573323 HM573324 HM573325 MH043953 MH043954 MK045992 MK045993 MK046000 MK046002 ABD18927.1 ACB30195.1 ACB30196.1 ADP21330.1 ADP21331.1 ADP21332.1 ADP21333.1 ADP21334.1 AVZ61108.1 AVZ61118.1 QBG67061.1 QBG67062.1 QBG67069.1 QBG67071.1 AF239308 AF239309 AF391541 MH203059 AXP11691.1 EF424615 ABP38240.1 DQ389648 ABD18924.1 MK045994 QBG67063.1 KF906249 AHN64773.1 EF424618 ABP38271.1 AH014871 AAY33943.1 EF424617 ABP38257.1 MH203062 AXP11694.1 EF424619 ABP38274.1 DQ682406 ABG78747.1 DQ389647 ABD18923.1 MK045998 MK045999 QBG67067.1 QBG67068.1 AH014866 AH014870 AAY33928.1 AAY33940.1 EF424616 ABP38246.1 DQ389649 ABD18925.1 MK046001 QBG67070.1 DQ994167 ABI98028.1 AF391542 DQ915164 ABI94000.1
MH687969 AYR18606.1 MH687973 MH687974 MH687977 MH687978 AYR18642.1 AYR18651.1 AYR18678.1 AYR18687.1 MH687971 MH687972 AYR18624.1 AYR18633.1 KY370047 ATP66749.1 KF294357 AID16630.1 MH687975 AYR18660.1 KY370052 ATP66778.1 KY370046 ATP66743.1 MN514966 QEY10656.1 KU886219 ANJ04973.1 MN514963 QEY10632.1 MH475914 QBH22432.1 MN514964 MN514965 QEY10640.1 QEY10648.1 MN514967 QEY10664.1 FJ938067 ACT11029.1 FJ415324 ACJ35485.1 U00735 S50936 M80842 DQ011855 AAY68296.1 MK095145 MK095146 AZS64192.1 AZS64193.1 GQ918142 JX860640 ACX46849.1 AFW97359.1 EU401979 ACB30199.1 GQ918143 ACX46859.1 L38962 L38963 U06093 AF239306 AF239307 AF230523 AF230524 AF230525 AF230526 AF230527 AF230528 AF339836 AF220295 S69264 M76372 FJ938063 ACT10982.1 DQ811784 ABG89283.1 MK095135 MK095136 MK095137 AZS64182.1 AZS64183.1 AZS64184.1 MK045995 QBG67064.1 MK095140 AZS64187.1 FJ425189 ACJ66999.1 FJ425186 ACJ66968.1 FJ425185 ACJ66956.1 JN874561 AFE48816.1 JN874559 AFE48794.1 FJ425184 ACJ66944.1 FJ425190 ACJ67011.1 GQ918141 ACX46839.1 MK095147 AZS64194.1 MH043952 AVZ61098.1 EU401977 EU401978 AB354579 ACB30197.1 ACB30198.1 BAF75630.1 MK095138 MK095139 AZS64185.1 AZS64186.1 MH203060 MH203061 MH203063 AXP11692.1 AXP11693.1 AXP11695.1 M84486 AF058942 KX432213 AQT26497.1 MN514962 QEY10624.1 FJ425188 ACJ66986.1 JN874560 AFE48804.1 MK095142 AZS64189.1 MG757138 AVI14999.1 DQ389643 DQ389644 DQ389645 ABD18919.1 ABD18920.1 ABD18921.1 KT368891 ALA50079.1 MF593476 ATI09448.1 EU999954 ACL12995.1 MH249786 QAY30019.1 MG757139 MG757141 AVI15010.1 AVI15032.1 AF058944 KU558922 ANJ04716.1 FJ938064 ACT10994.1 FJ938065 ACT11006.1 JN874562 AFE48826.1 EF424620 ABP38286.1 KX982264 ARD05998.1 EF445634 ABP73655.1 KF906250 KF906251 AHN64782.1 AHN64791.1 EU019216 ABV74053.1 MG518518 AVN88333.1 EF424624 ABP38338.1 EF424623 ABP38323.1 FJ425187 ACJ66975.1 EF424621 ABP38304.1 EF424622 ABP38312.1 MH043955 AVZ61128.1 DQ994168 ABI98029.1 DQ389651 EU401975 EU401976 HM573321 HM573322 HM573323 HM573324 HM573325 MH043953 MH043954 MK045992 MK045993 MK046000 MK046002 ABD18927.1 ACB30195.1 ACB30196.1 ADP21330.1 ADP21331.1 ADP21332.1 ADP21333.1 ADP21334.1 AVZ61108.1 AVZ61118.1 QBG67061.1 QBG67062.1 QBG67069.1 QBG67071.1 AF239308 AF239309 AF391541 MH203059 AXP11691.1 EF424615 ABP38240.1 DQ389648 ABD18924.1 MK045994 QBG67063.1 KF906249 AHN64773.1 EF424618 ABP38271.1 AH014871 AAY33943.1 EF424617 ABP38257.1 MH203062 AXP11694.1 EF424619 ABP38274.1 DQ682406 ABG78747.1 DQ389647 ABD18923.1 MK045998 MK045999 QBG67067.1 QBG67068.1 AH014866 AH014870 AAY33928.1 AAY33940.1 EF424616 ABP38246.1 DQ389649 ABD18925.1 MK046001 QBG67070.1 DQ994167 ABI98028.1 AF391542 DQ915164 ABI94000.1
Proteomes
UP000153628
UP000174571
UP000144342
UP000164707
UP000158296
UP000007554
+ More
UP000125924 UP000137675 UP000008572 UP000172225 UP000147981 UP000133443 UP000140344 UP000104294 UP000125269 UP000107676 UP000174569 UP000102696 UP000168103 UP000148210 UP000124585 UP000158231 UP000136745 UP000139966 UP000145001 UP000170940 UP000145507 UP000149260 UP000147178 UP000163913 UP000158327 UP000136116 UP000139235 UP000008570 UP000121953 UP000121258 UP000173415 UP000151273 UP000134650 UP000127099 UP000008571 UP000154814
UP000125924 UP000137675 UP000008572 UP000172225 UP000147981 UP000133443 UP000140344 UP000104294 UP000125269 UP000107676 UP000174569 UP000102696 UP000168103 UP000148210 UP000124585 UP000158231 UP000136745 UP000139966 UP000145001 UP000170940 UP000145507 UP000149260 UP000147178 UP000163913 UP000158327 UP000136116 UP000139235 UP000008570 UP000121953 UP000121258 UP000173415 UP000151273 UP000134650 UP000127099 UP000008571 UP000154814
Interpro
SUPFAM
SSF49818
SSF49818
ProteinModelPortal
A0A0A7UX90
A0A0A7V3Y4
A0A0A7V3Z7
A0A3G3NHC2
A0A3G3NH50
A0A3G3NHN3
+ More
A0A2H4MXC4 A0A096XNF9 A0A3G3NHC5 A0A2H4MX16 A0A2H4MZ34 A0A5J6NRP2 A0A191URZ4 A0A5J6NUD6 A0A481SAP1 A0A5J6NRK9 A0A5J6NRQ9 C6GHQ9 B7TYG8 P15776 Q66165 Q2QKN4 A0A451ETZ3 D0EY82 B9TXU2 D0EY92 P59709 P59710 C6GHL2 Q0PL40 A0A451ETZ0 A0A493R5L6 A0A3S9Y2C0 B7U2R4 B7U2N2 B7U2M1 H9AA54 H9AA33 B7U2L0 B7U2S6 D0EY72 A0A3S9Y2S0 A0A2R4STV0 A7BKC1 A0A3Q9K3J3 A0A346I7H2 P31613 A0A1S6KXR2 A0A5J6NRI4 B7U2Q3 H9AA43 A0A3S9Y2A6 A0A3S7GXV6 Q2EEZ7 A0A0U2GN17 A0A291L0R5 B8RIQ6 A0A411D561 A0A3S7GY11 Q9QAQ9 A0A191URB1 C6GHM4 C6GHN6 H9AA64 A4ZU29 A0A1V0JAY6 A6YD39 X2JFB2 A8HB15 A0A2P1IQB6 A4ZU73 A4ZU62 B7U2P3 A4ZU40 A4ZU51 A0A2R4STW8 A0A1J3 Q2EEZ0 Q9DR81 P59711 A0A346I7H1 A4ZTX4 Q2EEZ3 A0A481S1W3 X2JHM7 A4ZU07 Q4U3S2 A4ZTZ6 A0A346I7H4 A4ZU18 A3E2F1 Q2EEZ4 A0A481S1A3 Q4U3S3 A4ZTY5 Q2EEZ2 A0A481S2D5 A0A1J2 Q8V437 Q06BD8
A0A2H4MXC4 A0A096XNF9 A0A3G3NHC5 A0A2H4MX16 A0A2H4MZ34 A0A5J6NRP2 A0A191URZ4 A0A5J6NUD6 A0A481SAP1 A0A5J6NRK9 A0A5J6NRQ9 C6GHQ9 B7TYG8 P15776 Q66165 Q2QKN4 A0A451ETZ3 D0EY82 B9TXU2 D0EY92 P59709 P59710 C6GHL2 Q0PL40 A0A451ETZ0 A0A493R5L6 A0A3S9Y2C0 B7U2R4 B7U2N2 B7U2M1 H9AA54 H9AA33 B7U2L0 B7U2S6 D0EY72 A0A3S9Y2S0 A0A2R4STV0 A7BKC1 A0A3Q9K3J3 A0A346I7H2 P31613 A0A1S6KXR2 A0A5J6NRI4 B7U2Q3 H9AA43 A0A3S9Y2A6 A0A3S7GXV6 Q2EEZ7 A0A0U2GN17 A0A291L0R5 B8RIQ6 A0A411D561 A0A3S7GY11 Q9QAQ9 A0A191URB1 C6GHM4 C6GHN6 H9AA64 A4ZU29 A0A1V0JAY6 A6YD39 X2JFB2 A8HB15 A0A2P1IQB6 A4ZU73 A4ZU62 B7U2P3 A4ZU40 A4ZU51 A0A2R4STW8 A0A1J3 Q2EEZ0 Q9DR81 P59711 A0A346I7H1 A4ZTX4 Q2EEZ3 A0A481S1W3 X2JHM7 A4ZU07 Q4U3S2 A4ZTZ6 A0A346I7H4 A4ZU18 A3E2F1 Q2EEZ4 A0A481S1A3 Q4U3S3 A4ZTY5 Q2EEZ2 A0A481S2D5 A0A1J2 Q8V437 Q06BD8
PDB
3CL4
E-value=2.90633e-146,
Score=1330
Ontologies
GO
GO:0016021 C:integral component of membrane
GO:0046789 F:host cell surface receptor binding
GO:0019031 C:viral envelope
GO:0001681 F:sialate O-acetylesterase activity
GO:0020002 C:host cell plasma membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0055036 C:virion membrane
GO:0005102 F:signaling receptor binding
GO:0042803 F:protein homodimerization activity
GO:0042802 F:identical protein binding
GO:0002683 P:negative regulation of immune system process
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0046789 F:host cell surface receptor binding
GO:0019031 C:viral envelope
GO:0001681 F:sialate O-acetylesterase activity
GO:0020002 C:host cell plasma membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0055036 C:virion membrane
GO:0005102 F:signaling receptor binding
GO:0042803 F:protein homodimerization activity
GO:0042802 F:identical protein binding
GO:0002683 P:negative regulation of immune system process
GO:0046813 P:receptor-mediated virion attachment to host cell
Subcellular Location
From MSLVP
Single-Pass Membrane
From Uniprot
Virion membrane
In infected cells becomes incorporated into the envelope of virions during virus assembly at the endoplasmic reticulum and cis Golgi. However, some may escape incorporation into virions and subsequently migrate to the cell surface. With evidence from 1 publications.
Host cell membrane In infected cells becomes incorporated into the envelope of virions during virus assembly at the endoplasmic reticulum and cis Golgi. However, some may escape incorporation into virions and subsequently migrate to the cell surface. With evidence from 1 publications.
Host cell membrane In infected cells becomes incorporated into the envelope of virions during virus assembly at the endoplasmic reticulum and cis Golgi. However, some may escape incorporation into virions and subsequently migrate to the cell surface. With evidence from 1 publications.
Topology
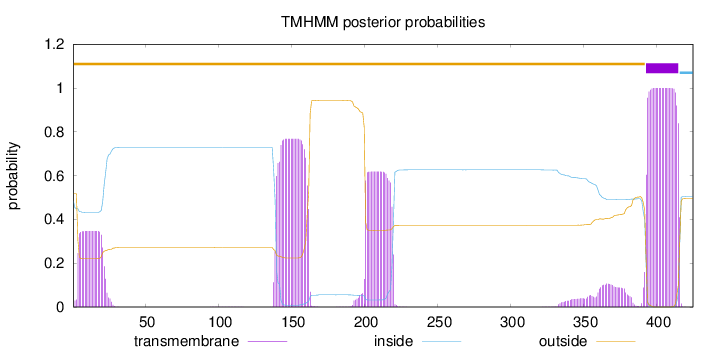
Length:
425
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
61.73407
Exp number, first 60 AAs:
6.59193
Total prob of N-in:
0.48232
outside
1 - 392
TMhelix
393 - 415
inside
416 - 425
Population Genetic Test Statistics
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 |
Protein |
AAT84353.1 |
|
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 |
Protein |
YP_009113024.1 |
|
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.