Strain
ARABIA_CAMEL_2015_NC_028752
(Region: Saudi Arabia; Strain: Camel alphacoronavirus isolate camel/Riyadh/Ry141/2015, complete genome.; Date: 2015)
Gene
spike protein
Description
Annotated in NCBI,
spike protein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGTTTGTACTTGTTGCATATGCCTTGTTGCATATTGCTGGTTGTCAAACTATAAATGTTCTGAACACTAGTCACTCTGTTTGTAACGGCTGTGTTGGTTATTCAGAAAATGTATTTGCTGTTGAGAGTGGTGGTTATATACCCTCCGACTTTGCATTTAATAATTGGTTTCTCTTAACTAATACCTCATCTGTTGTAGACGGTGTTGTGAGGAGTTTTCAGCCTTTGTTGCTTAATTGCTTATGGTCTGTTTCTGGYTCGCGGTTTACTACTGGTTTTGTCTATTTTAATGGTACTGGGAGAGGTGATTGTAAAGGTTTTTCTTCAAATGTTTTGTCTGATGTCATACGTTACAACATCAATTTTGAAGAAAACCTTAGACGTGGAACCATTTTGTTTAAAACATCTTTTGGTGTTGTTGTGTTTTATTGTACCAACAACACTTTAGTTTCAGGTGATGCTTACATACCATTTGGTACAGTTTTGGGCAATTTTTATTGCTTTGTAAATACTACTATTGGCAATGAAACTACTTCTTCTTTTGTGGGTGCACTACCTAAGACAGTTCGTGAGTTTGTTATTTCACGCACAGGACATTTTTATATTAATGGCTATCGCTATTTTACTTTAGGTAATTTAGAAGCCGTTAATTTCAATGTCACTAATGCAGAAACCACTTTTTGTACTGTTGCGTTAGCTTCTTATGCTGACGTTTTGGTTAATGTGTCACAAACCGCTATTGCTAATATAATTTATTGCAACTCTGTTATTAACAGACTGAGATGTGACCAGTTGTCCTTTGATGTACCAGATGGTTTTTATTCCACAAGTCCTATTCAATCCGTTGAGCTACCTGTGTCTATTGTGTCGCTACCTGTTTATCACAAACATACGTTTATTGTGTTGTATGTTGATTTTAAACCTCAGAGTGGCGCCGGCACGTGTTATAACTGTCGTCCTTCTGTTGTTAATATTACACTGGCCAATTTTAATGAAACTAAAGGGCCTTTGTGTGTTGAAACATCACACTTCACTACCAAATACGTTGCTTCCAATGTTGGTAGGTGGAGTGCTAGTATTAACACGGGAAATTGCCCTTTTTCTTTTGGCAAAGTTAATAACTTTGTTAAATTTGGCAGTGTATGTTTTTCACTAAAGGATATACCCGGTGGTTGCGCAATGCCTATAGTGGCTAATTTTGCTTATATTAATTCTTATACTATAGGCTCATTGTATGTTTCTTGGAGTGATGGTGATGGAATTACTGGCGTCCCAAAACCTGTTGAGGGTGTTAGTTCCTTTATGAATGTTACATTGAATAAATGTACTAAATATAATATTTATGATGTATCTGGTGTGGGTGTTATTCGCATTAGCAATGACACCTTTCTTAATGGAATTACGTACACATCAACTTCAGGTAACCTTTTGGGTTTCAAAGATGTTACTAATGGTATCATCTACTCTATCACTCCTTGTAACCCACCAGATCAGCTTGTTGTTTATCAGCAAGCTGTTGTTGGTGCTATGTTGTCTGAAAATTCAAGTAGTTACGGCTTCTCTAATGTTGTGGAATTGCCTAACTTTTTCTACGCTTCTAATGGCACTTACAATTGCACAGATGCTGTTTTAACTTATTCTAGCTTTGGTGTTTGTGCAGATGGTTCTATAATTGCTGTTCAACCACGCAATGTTTCTTATGATGGTGTCTCAGCGATTGTCACAGCTAATTTGTCTATACCTTCCAATTGGACTACTTCAGTCCAGGTTGAGTATTTACAAATTACAAGCACACCTATTTTAGTGGATTGCTCCACTTATGTTTGCAATGGTAATGTGCGCTGTGTTGAATTGCTTAAGCAGTATACTTCTGCTTGTAAAACTATTGAAGATGCCTTAAGAATTAGTGCTATGCTGGAGTCTGCAGATGTTGGTGAGATGCTCACTTTTGACGAGAAAGCGTTTACACTTGCTAATGTTAGTAGTTTTGGTGACTACAACCTTAGCAGTGTTATACCTAGCTTGCCTACAAGTGGTAGTAGAGTAGCCGGTCGCAGTGCCATAGAAGACATACTTTTTAGTAAAGTTGTTACTTCTGGACTTGGCACTGTGGACGCAGACTACAAAAAGTGCACTAAGGGTCTTTCCATTGCTGATTTGGCTTGTGCTCAATATTATAATGGCATTATGGTTTTGCCTGGCGTTGCTGATGCTGAACGAATGGCCATGTACACAGGTTCCTTAATTGGTGGCATTGCCTTAGGAGGTCTTACATCAGCCGCTGCAATACCATTTTCTTTAGCTCTTCAGGCACGTTTGAACTATGTGGCATTGCAGACTGATGTTTTGCAAGAAAATCAGAAAATTCTTGCTGCATCTTTTAATAAAGCAATGACTAACATTGTCGATGCCTTTACAGGAGTTAATGATGCTATTACTCAAACTTCACAAGCTATACAAACAGTTGCCACTGCACTTAATAAGATTCAGGATGTTGTGAATCAACAAGGCAATGCATTGAACCATCTAACTTCTCAGTTGAGGCAAAATTTTCAAGCTATTTCCAGTTCTATTCAGGCTATCTATGACAGACTTGACACTATTCAGGCTGATCAACAAGTAGATAGGCTTATTACTGGTAGACTGGCTGCTTTGAACGCATTTGTTGCTCAGACACTTACTAAGTATACTGAGGTGCGTGCTTCTAGACAGCTTGCACAACAAAAAGTGAATGAATGTGTCAAATCCCAGTCTAACCGTTATGGTTTCTGTGGAAATGGCACACACATTTTCTCAATTGTGAATTCTGCTCCTGAAGGCCTTGTTTTTCTTCACACTGTCTTGTTGCCCACGCAATATAAGGATGTTGAGGCGTGGTCCGGGCTTTGCGTTGATAATACAAACGGTTATGTGCTGCGACAACCTAATCTTGCTCTTTACAAAGAAGGCAATTATTATAGAATCACATCACGTTTTATGTTTGAACCACGCATTCCTACTATGGCAGATTTTGTCCAGATTGAAAATTGCAATGTCACATTTGTTAACATTTCTCGCTCTGAGTTGCAAACTATTGTGCCAGAGTATATTGATGTTAACAAGACGCTGCAAGAATTAATTGACAAATTGCCAAATTATACTGTCCCAGACCTAGGCATCGACCAGTACAACCAGACTATTTTGAATTTGACCAGTGAAATTAGCACCCTTGAAAATAAATCTGCGGAGCTTAATTACACTGTTCAAAGACTGCAAACTCTTATTGACAACATAAATAGCACTTTAGTCGACTTAAAGTGGCTCAACCGGGTTGAGACTTATCTCAAGTGGCCGTGGTGGGTGTGGTTGTGCATTTCAGTCGTGCTCATCTTTGTGGTGAGTGTTTTGCTATTATGTTGTTGCTCTACTGGTTGCTGTGGCTTCTTTAGTTGTCTTGCATCTTCTACTAGAGGTTGTTGTGAGTCAACTAAACTTCCTTATTACGACGTTGAAAAGATTCACATACAGTAA
Protein
MFVLVAYALLHIAGCQTINVLNTSHSVCNGCVGYSENVFAVESGGYIPSDFAFNNWFLLTNTSSVVDGVVRSFQPLLLNCLWSVSGSRFTTGFVYFNGTGRGDCKGFSSNVLSDVIRYNINFEENLRRGTILFKTSFGVVVFYCTNNTLVSGDAYIPFGTVLGNFYCFVNTTIGNETTSSFVGALPKTVREFVISRTGHFYINGYRYFTLGNLEAVNFNVTNAETTFCTVALASYADVLVNVSQTAIANIIYCNSVINRLRCDQLSFDVPDGFYSTSPIQSVELPVSIVSLPVYHKHTFIVLYVDFKPQSGAGTCYNCRPSVVNITLANFNETKGPLCVETSHFTTKYVASNVGRWSASINTGNCPFSFGKVNNFVKFGSVCFSLKDIPGGCAMPIVANFAYINSYTIGSLYVSWSDGDGITGVPKPVEGVSSFMNVTLNKCTKYNIYDVSGVGVIRISNDTFLNGITYTSTSGNLLGFKDVTNGIIYSITPCNPPDQLVVYQQAVVGAMLSENSSSYGFSNVVELPNFFYASNGTYNCTDAVLTYSSFGVCADGSIIAVQPRNVSYDGVSAIVTANLSIPSNWTTSVQVEYLQITSTPILVDCSTYVCNGNVRCVELLKQYTSACKTIEDALRISAMLESADVGEMLTFDEKAFTLANVSSFGDYNLSSVIPSLPTSGSRVAGRSAIEDILFSKVVTSGLGTVDADYKKCTKGLSIADLACAQYYNGIMVLPGVADAERMAMYTGSLIGGIALGGLTSAAAIPFSLALQARLNYVALQTDVLQENQKILAASFNKAMTNIVDAFTGVNDAITQTSQAIQTVATALNKIQDVVNQQGNALNHLTSQLRQNFQAISSSIQAIYDRLDTIQADQQVDRLITGRLAALNAFVAQTLTKYTEVRASRQLAQQKVNECVKSQSNRYGFCGNGTHIFSIVNSAPEGLVFLHTVLLPTQYKDVEAWSGLCVDNTNGYVLRQPNLALYKEGNYYRITSRFMFEPRIPTMADFVQIENCNVTFVNISRSELQTIVPEYIDVNKTLQELIDKLPNYTVPDLGIDQYNQTILNLTSEISTLENKSAELNYTVQRLQTLIDNINSTLVDLKWLNRVETYLKWPWWVWLCISVVLIFVVSVLLLCCCSTGCCGFFSCLASSTRGCCESTKLPYYDVEKIHIQ
Summary
Function
S1 region attaches the virion to the cell membrane by interacting with host ANPEP/aminopeptidase N, initiating the infection. Binding to the receptor probably induces conformational changes in the S glycoprotein unmasking the fusion peptide of S2 region and activating membranes fusion. S2 region belongs to the class I viral fusion protein. Under the current model, the protein has at least 3 conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) regions assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer. During virus morphogenesis, found in a complex with M and HE proteins. Interacts with host ANPEP.
Similarity
Belongs to the alphacoronaviruses spike protein family.
Keywords
3D-structure
Coiled coil
Glycoprotein
Host membrane
Host-virus interaction
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Virion
Virulence
Virus entry into host cell
Feature
chain Spike glycoprotein
Uniprot
A0A0U2GRB2
A0A0U2GRL6
A0A0U2GQ76
A0A0U2GN50
A0A0U2GNG3
A0A0U2GMN8
+ More
A0A0U2GMQ4 I1VWF5 A0A1C8YZ37 A0A1C8YZ34 A0A0U2GME6 A0A1C8YZ26 A0A0U2GQI5 A0A0U2GMG2 A0A0U2GN82 A0A1B3TNA4 Q1HVM5 Q1HVM3 Q1HVL9 Q1HVM0 Q1HVM1 Q1HVL7 Q1HVL8 P15423 S5YNL7 Q1HVM6 H1AG29 A0A059SGA9 S5YGU2 Q1HVL4 Q1HVL3 S5YAI3 S5YGT1 S5YNN4 Q1HVK9 A0A384RK44 Q1HVK6 H1AG30 Q1HVK5 J9VBQ8 A0A1V0E0W8 Q1HVL0 A0A384QXY4 Q1HVK3 Q1HVL1 Q1HVK4 A0A384QXM1 H1AG31 A0A384QXM2 H1AG33 J9UWK8 A0A384RJX8 A0A384RBT6 A0A1Z1R0Z6 A0A384RNQ9 A0A384R2L9 A0A384RJY2 A0A384RBT3 A0A384RJY8 A0A2S1PV28 H1AG32 A0A384QXL8 A0A384R9G2 A0A384RBT4 A0A384RK26 A0A1Y0EV30 A0A384RNP7 A0A1B3TN99 A0A1L7B942 A0A384RHP9 A0A1L7B8Z9 A0A1W6DZS3 A0A1L7B908 A0A384R9G0 A0A1L7B931 A0A1L7B944 A0A1L7B934 A0A1V0BZ86 A0A5B9MYJ0 A0A5C2D4L1 A0A223FUI6 A0A0P0KI80 A0A0P0KR98 A0A0N7J129 A0A0P0K0X5 A0A0P0KBC5 A0A0N7J128 A0A0P0KXK6 A0A1L2KGF1 A0A1L2KGE4 A0A0P0K6L9 A0A0P0KMM1 A0A0P0KR90 A0A0P0KRB3 A0A0P0KWD4 A0A0P0KH07 Q06Y16 A0A0K1D6F6 A0A513ZT30 A0A1V0E0U5
A0A0U2GMQ4 I1VWF5 A0A1C8YZ37 A0A1C8YZ34 A0A0U2GME6 A0A1C8YZ26 A0A0U2GQI5 A0A0U2GMG2 A0A0U2GN82 A0A1B3TNA4 Q1HVM5 Q1HVM3 Q1HVL9 Q1HVM0 Q1HVM1 Q1HVL7 Q1HVL8 P15423 S5YNL7 Q1HVM6 H1AG29 A0A059SGA9 S5YGU2 Q1HVL4 Q1HVL3 S5YAI3 S5YGT1 S5YNN4 Q1HVK9 A0A384RK44 Q1HVK6 H1AG30 Q1HVK5 J9VBQ8 A0A1V0E0W8 Q1HVL0 A0A384QXY4 Q1HVK3 Q1HVL1 Q1HVK4 A0A384QXM1 H1AG31 A0A384QXM2 H1AG33 J9UWK8 A0A384RJX8 A0A384RBT6 A0A1Z1R0Z6 A0A384RNQ9 A0A384R2L9 A0A384RJY2 A0A384RBT3 A0A384RJY8 A0A2S1PV28 H1AG32 A0A384QXL8 A0A384R9G2 A0A384RBT4 A0A384RK26 A0A1Y0EV30 A0A384RNP7 A0A1B3TN99 A0A1L7B942 A0A384RHP9 A0A1L7B8Z9 A0A1W6DZS3 A0A1L7B908 A0A384R9G0 A0A1L7B931 A0A1L7B944 A0A1L7B934 A0A1V0BZ86 A0A5B9MYJ0 A0A5C2D4L1 A0A223FUI6 A0A0P0KI80 A0A0P0KR98 A0A0N7J129 A0A0P0K0X5 A0A0P0KBC5 A0A0N7J128 A0A0P0KXK6 A0A1L2KGF1 A0A1L2KGE4 A0A0P0K6L9 A0A0P0KMM1 A0A0P0KR90 A0A0P0KRB3 A0A0P0KWD4 A0A0P0KH07 Q06Y16 A0A0K1D6F6 A0A513ZT30 A0A1V0E0U5
Pubmed
EMBL
KT368907
KT368908
ALA50193.1
ALA50200.1
KT368903
ALA50165.1
+ More
KT368899 ALA50137.1 KT368898 ALA50130.1 KT368916 ALA50256.1 KT368892 KT368893 KT368894 KT368902 KT368904 KT368905 KT368906 KT368909 KT368910 KT368911 KT368912 KT368913 KT368914 ALA50088.1 ALA50095.1 ALA50102.1 ALA50158.1 ALA50172.1 ALA50179.1 ALA50186.1 ALA50207.1 ALA50214.1 ALA50221.1 ALA50228.1 ALA50235.1 ALA50242.1 KT368901 ALA50151.1 JQ410000 AFI49431.1 KT253324 KT253326 KT253328 AOI28259.1 AOI28275.1 AOI28291.1 KT253327 AOI28283.1 KT368895 KT368896 MF593473 ALA50109.1 ALA50116.1 ATI09437.1 KT253325 AOI28267.1 KT368915 ALA50249.1 KT368897 ALA50123.1 KT368900 ALA50144.1 KU291449 AOG74789.1 DQ243964 ABB90507.1 DQ243966 ABB90509.1 DQ243970 ABB90513.1 DQ243965 DQ243969 ABB90508.1 ABB90512.1 DQ243967 DQ243968 ABB90510.1 ABB90511.1 DQ243972 ABB90515.1 DQ243971 ABB90514.1 X16816 AF304460 AF344186 AF344187 AF344188 AF344189 Y09923 Y10051 Y10052 X15654 KF514429 AGT21338.1 DQ243963 ABB90506.1 AB691763 BAL45637.1 KF293664 KF293665 KF293666 AGW80948.1 KF514432 AGT21360.1 DQ243973 DQ243974 DQ243975 ABB90516.1 ABB90517.1 ABB90518.1 DQ243976 ABB90519.1 KF514433 AGT21367.1 KF514430 AGT21345.1 KF514431 AGT21353.1 DQ243980 ABB90523.1 KM055559 AIW52756.1 DQ243983 ABB90526.1 AB691764 BAL45638.1 DQ243984 ABB90527.1 JX503061 AFR79257.1 KY674919 ARB07425.1 DQ243979 ABB90522.1 KM055557 AIW52754.1 DQ243986 ABB90529.1 DQ243977 DQ243978 ABB90520.1 ABB90521.1 DQ243985 ABB90528.1 KM055534 KM055545 KM055546 KM055547 AIW52731.1 AIW52742.1 AIW52743.1 AIW52744.1 AB691765 BAL45639.1 KM055555 AIW52752.1 KM055552 KM055554 AB691767 AIW52749.1 AIW52751.1 BAL45641.1 JX503060 AFR79250.1 KM055532 KM055533 AIW52729.1 AIW52730.1 KM055558 AIW52755.1 KY996417 ARX11396.1 KM055551 AIW52748.1 KM055556 AIW52753.1 KM055543 AIW52740.1 KM055538 AIW52735.1 KM055553 AIW52750.1 MH048989 AWH62679.1 AB691766 BAL45640.1 KM055535 KM055536 KM055537 KM055539 KM055540 KM055541 KM055542 AIW52732.1 AIW52733.1 AIW52734.1 AIW52736.1 AIW52737.1 AIW52738.1 AIW52739.1 KM055560 AIW52757.1 KM055548 AIW52745.1 KM055549 AIW52746.1 KY983587 ARU07601.1 KM055531 AIW52728.1 KU291448 AOG74783.1 KY369913 APT69883.1 KM055544 AIW52741.1 KY369908 APT69849.1 KY967357 ARK08642.1 KY369909 KY674914 KY621348 APT69856.1 ARB07392.1 ARK08620.1 KM055550 AIW52747.1 KY369912 APT69876.1 KY369914 APT69890.1 KY369910 KY369911 APT69862.1 APT69869.1 KY684760 ARA15429.1 MN306046 QEG03785.1 MN369046 QEO75985.1 MF542265 AST12964.1 KT253266 ALK28772.1 KT253270 ALK28781.1 KT253269 ALK28775.1 KT253265 ALK28771.1 KT253267 ALK28773.1 KT253261 KT253263 ALK28767.1 ALK28769.1 KT253262 ALK28768.1 KY073748 APD51507.1 KY073747 APD51499.1 KT253271 ALK28787.1 KT253268 ALK28774.1 KT253264 ALK28770.1 KT253272 ALK28793.1 KT253259 ALK28765.1 KT253260 ALK28766.1 DQ445911 ABE97137.1 KT266906 KX179494 KX179495 KX179496 AKT07952.1 APF29061.1 APF29062.1 APF29063.1 MN026166 QDH43736.1 KY674916 ARB07406.1
KT368899 ALA50137.1 KT368898 ALA50130.1 KT368916 ALA50256.1 KT368892 KT368893 KT368894 KT368902 KT368904 KT368905 KT368906 KT368909 KT368910 KT368911 KT368912 KT368913 KT368914 ALA50088.1 ALA50095.1 ALA50102.1 ALA50158.1 ALA50172.1 ALA50179.1 ALA50186.1 ALA50207.1 ALA50214.1 ALA50221.1 ALA50228.1 ALA50235.1 ALA50242.1 KT368901 ALA50151.1 JQ410000 AFI49431.1 KT253324 KT253326 KT253328 AOI28259.1 AOI28275.1 AOI28291.1 KT253327 AOI28283.1 KT368895 KT368896 MF593473 ALA50109.1 ALA50116.1 ATI09437.1 KT253325 AOI28267.1 KT368915 ALA50249.1 KT368897 ALA50123.1 KT368900 ALA50144.1 KU291449 AOG74789.1 DQ243964 ABB90507.1 DQ243966 ABB90509.1 DQ243970 ABB90513.1 DQ243965 DQ243969 ABB90508.1 ABB90512.1 DQ243967 DQ243968 ABB90510.1 ABB90511.1 DQ243972 ABB90515.1 DQ243971 ABB90514.1 X16816 AF304460 AF344186 AF344187 AF344188 AF344189 Y09923 Y10051 Y10052 X15654 KF514429 AGT21338.1 DQ243963 ABB90506.1 AB691763 BAL45637.1 KF293664 KF293665 KF293666 AGW80948.1 KF514432 AGT21360.1 DQ243973 DQ243974 DQ243975 ABB90516.1 ABB90517.1 ABB90518.1 DQ243976 ABB90519.1 KF514433 AGT21367.1 KF514430 AGT21345.1 KF514431 AGT21353.1 DQ243980 ABB90523.1 KM055559 AIW52756.1 DQ243983 ABB90526.1 AB691764 BAL45638.1 DQ243984 ABB90527.1 JX503061 AFR79257.1 KY674919 ARB07425.1 DQ243979 ABB90522.1 KM055557 AIW52754.1 DQ243986 ABB90529.1 DQ243977 DQ243978 ABB90520.1 ABB90521.1 DQ243985 ABB90528.1 KM055534 KM055545 KM055546 KM055547 AIW52731.1 AIW52742.1 AIW52743.1 AIW52744.1 AB691765 BAL45639.1 KM055555 AIW52752.1 KM055552 KM055554 AB691767 AIW52749.1 AIW52751.1 BAL45641.1 JX503060 AFR79250.1 KM055532 KM055533 AIW52729.1 AIW52730.1 KM055558 AIW52755.1 KY996417 ARX11396.1 KM055551 AIW52748.1 KM055556 AIW52753.1 KM055543 AIW52740.1 KM055538 AIW52735.1 KM055553 AIW52750.1 MH048989 AWH62679.1 AB691766 BAL45640.1 KM055535 KM055536 KM055537 KM055539 KM055540 KM055541 KM055542 AIW52732.1 AIW52733.1 AIW52734.1 AIW52736.1 AIW52737.1 AIW52738.1 AIW52739.1 KM055560 AIW52757.1 KM055548 AIW52745.1 KM055549 AIW52746.1 KY983587 ARU07601.1 KM055531 AIW52728.1 KU291448 AOG74783.1 KY369913 APT69883.1 KM055544 AIW52741.1 KY369908 APT69849.1 KY967357 ARK08642.1 KY369909 KY674914 KY621348 APT69856.1 ARB07392.1 ARK08620.1 KM055550 AIW52747.1 KY369912 APT69876.1 KY369914 APT69890.1 KY369910 KY369911 APT69862.1 APT69869.1 KY684760 ARA15429.1 MN306046 QEG03785.1 MN369046 QEO75985.1 MF542265 AST12964.1 KT253266 ALK28772.1 KT253270 ALK28781.1 KT253269 ALK28775.1 KT253265 ALK28771.1 KT253267 ALK28773.1 KT253261 KT253263 ALK28767.1 ALK28769.1 KT253262 ALK28768.1 KY073748 APD51507.1 KY073747 APD51499.1 KT253271 ALK28787.1 KT253268 ALK28774.1 KT253264 ALK28770.1 KT253272 ALK28793.1 KT253259 ALK28765.1 KT253260 ALK28766.1 DQ445911 ABE97137.1 KT266906 KX179494 KX179495 KX179496 AKT07952.1 APF29061.1 APF29062.1 APF29063.1 MN026166 QDH43736.1 KY674916 ARB07406.1
Proteomes
UP000120649
UP000151210
UP000103783
UP000136947
UP000116691
UP000152663
+ More
UP000100137 UP000103248 UP000108758 UP000110675 UP000115199 UP000115399 UP000124352 UP000128900 UP000136940 UP000152773 UP000153580 UP000171347 UP000173795 UP000173458 UP000146121 UP000101679 UP000169049 UP000153625 UP000147133 UP000104585 UP000006716 UP000110267 UP000139984 UP000137891 UP000143565 UP000105041 UP000122373 UP000118770 UP000096398 UP000130985 UP000115484 UP000141634
UP000100137 UP000103248 UP000108758 UP000110675 UP000115199 UP000115399 UP000124352 UP000128900 UP000136940 UP000152773 UP000153580 UP000171347 UP000173795 UP000173458 UP000146121 UP000101679 UP000169049 UP000153625 UP000147133 UP000104585 UP000006716 UP000110267 UP000139984 UP000137891 UP000143565 UP000105041 UP000122373 UP000118770 UP000096398 UP000130985 UP000115484 UP000141634
PRIDE
ProteinModelPortal
A0A0U2GRB2
A0A0U2GRL6
A0A0U2GQ76
A0A0U2GN50
A0A0U2GNG3
A0A0U2GMN8
+ More
A0A0U2GMQ4 I1VWF5 A0A1C8YZ37 A0A1C8YZ34 A0A0U2GME6 A0A1C8YZ26 A0A0U2GQI5 A0A0U2GMG2 A0A0U2GN82 A0A1B3TNA4 Q1HVM5 Q1HVM3 Q1HVL9 Q1HVM0 Q1HVM1 Q1HVL7 Q1HVL8 P15423 S5YNL7 Q1HVM6 H1AG29 A0A059SGA9 S5YGU2 Q1HVL4 Q1HVL3 S5YAI3 S5YGT1 S5YNN4 Q1HVK9 A0A384RK44 Q1HVK6 H1AG30 Q1HVK5 J9VBQ8 A0A1V0E0W8 Q1HVL0 A0A384QXY4 Q1HVK3 Q1HVL1 Q1HVK4 A0A384QXM1 H1AG31 A0A384QXM2 H1AG33 J9UWK8 A0A384RJX8 A0A384RBT6 A0A1Z1R0Z6 A0A384RNQ9 A0A384R2L9 A0A384RJY2 A0A384RBT3 A0A384RJY8 A0A2S1PV28 H1AG32 A0A384QXL8 A0A384R9G2 A0A384RBT4 A0A384RK26 A0A1Y0EV30 A0A384RNP7 A0A1B3TN99 A0A1L7B942 A0A384RHP9 A0A1L7B8Z9 A0A1W6DZS3 A0A1L7B908 A0A384R9G0 A0A1L7B931 A0A1L7B944 A0A1L7B934 A0A1V0BZ86 A0A5B9MYJ0 A0A5C2D4L1 A0A223FUI6 A0A0P0KI80 A0A0P0KR98 A0A0N7J129 A0A0P0K0X5 A0A0P0KBC5 A0A0N7J128 A0A0P0KXK6 A0A1L2KGF1 A0A1L2KGE4 A0A0P0K6L9 A0A0P0KMM1 A0A0P0KR90 A0A0P0KRB3 A0A0P0KWD4 A0A0P0KH07 Q06Y16 A0A0K1D6F6 A0A513ZT30 A0A1V0E0U5
A0A0U2GMQ4 I1VWF5 A0A1C8YZ37 A0A1C8YZ34 A0A0U2GME6 A0A1C8YZ26 A0A0U2GQI5 A0A0U2GMG2 A0A0U2GN82 A0A1B3TNA4 Q1HVM5 Q1HVM3 Q1HVL9 Q1HVM0 Q1HVM1 Q1HVL7 Q1HVL8 P15423 S5YNL7 Q1HVM6 H1AG29 A0A059SGA9 S5YGU2 Q1HVL4 Q1HVL3 S5YAI3 S5YGT1 S5YNN4 Q1HVK9 A0A384RK44 Q1HVK6 H1AG30 Q1HVK5 J9VBQ8 A0A1V0E0W8 Q1HVL0 A0A384QXY4 Q1HVK3 Q1HVL1 Q1HVK4 A0A384QXM1 H1AG31 A0A384QXM2 H1AG33 J9UWK8 A0A384RJX8 A0A384RBT6 A0A1Z1R0Z6 A0A384RNQ9 A0A384R2L9 A0A384RJY2 A0A384RBT3 A0A384RJY8 A0A2S1PV28 H1AG32 A0A384QXL8 A0A384R9G2 A0A384RBT4 A0A384RK26 A0A1Y0EV30 A0A384RNP7 A0A1B3TN99 A0A1L7B942 A0A384RHP9 A0A1L7B8Z9 A0A1W6DZS3 A0A1L7B908 A0A384R9G0 A0A1L7B931 A0A1L7B944 A0A1L7B934 A0A1V0BZ86 A0A5B9MYJ0 A0A5C2D4L1 A0A223FUI6 A0A0P0KI80 A0A0P0KR98 A0A0N7J129 A0A0P0K0X5 A0A0P0KBC5 A0A0N7J128 A0A0P0KXK6 A0A1L2KGF1 A0A1L2KGE4 A0A0P0K6L9 A0A0P0KMM1 A0A0P0KR90 A0A0P0KRB3 A0A0P0KWD4 A0A0P0KH07 Q06Y16 A0A0K1D6F6 A0A513ZT30 A0A1V0E0U5
PDB
6U7H
E-value=0,
Score=5483
Ontologies
KEGG
GO
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0019031 C:viral envelope
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0016021 C:integral component of membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0019031 C:viral envelope
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0016021 C:integral component of membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
Subcellular Location
From MSLVP
Host nucleus. Host cytoplasm.
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. With evidence from 1 publications.
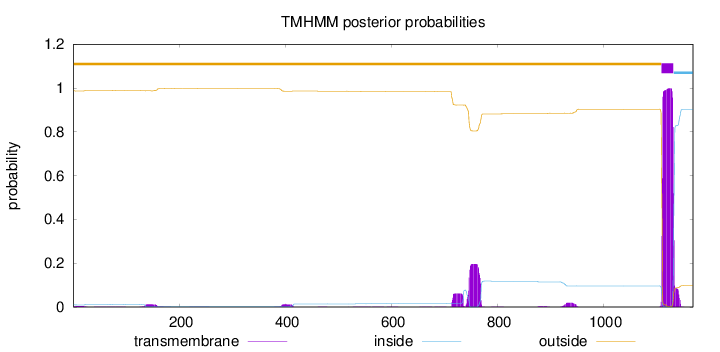
Topology
Length:
1169
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
30.16746
Exp number, first 60 AAs:
0.09741
Total prob of N-in:
0.01230
outside
1 - 1109
TMhelix
1110 - 1132
inside
1133 - 1169
Population Genetic Test Statistics
Pi
0.4167114
Theta
0.6102617
Tajima's D
-1.2794173
CLR
10.696814
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 |
Protein |
NP_073551.1 |
|
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 |
Protein |
YP_009194639.1 |
|
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.