Strain
UGANDA_BAT_2013_NC_034440
(Region: Uganda; Strain: Bat coronavirus isolate PREDICT/PDF-2180, complete genome.; Date: 2013)
Gene
spike glycoprotein
Description
Annotated in NCBI,
spike glycoprotein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGACGTACTCAGTGTCTCTACTGATGTGCTTGTTAACTTTTATAGGGGCAAACGCTAAAATTGTGTCGATACCTGGTGGTGTCGGTACTGGAGCTTGTCCCCAGGTTGATATGCAGCCCAGTTATTTTATAAAGCATAACTGGCCTGAACCTATTGACATGAATAAGGCAGACGGTGTCATCTACCCAAATGGCCGCACTTATTCTAACATCACATTACAGACCACTAATCTGTTTCCTCGTAATGGTGATTTAGGCACTCAGTATGTCTATTCAGCATCTAATGAGAAAAGCCGCACTAGCAATGTGGCTTTTATTAGTAATTATTCATACTATGGCAATCCCTTTGGCGACGGCATTGTCATACGTATAGGTCAAAATTCTAATAAGACTGGTAGTGTCATTGTGGGCACAGCACAGACTACTATTAAAAAGATCTACCCAGCTCTTATGCTTGGTAGTTCTTTTGGCAATTTCTCTGTTAATAATAAGTCAGGTGCTTATTTTAATCACACCCTTCTTATCTTACCTAGCAAGTGTGGCACTGTATTTCAGGTGGCATACTGCCTTCTACAACCAAGGACTGACTCTTATTGTCCCGGTAATGCTAACTATGTTAGCTATGCACTCATTGATTCTCCTACGGATTGTACATCTGCGGATGAATCTAAACGTAGGAATGGTCTTGAGGACATTAAAAAGTACTTCAATTTGGTCAATTGTACCTATTTTGAAGAGTTTAATGTCACAGCTGACGAGCGTGCAGAGTGGTTTGGCATCACCCAAGATTCTCAGGGTGTGCACCTCTACACCTCTCGTAAAAATGGTTTCAATTCAAATAATCTCTTTTTATTTGCTTCTGTGCCCATTTATGATAAGATCAATTACTACACTGTAATTCCTCGCTCAATAATAACTCCTGCCAACCAGCGTTCTGCTTGGGCAGCATTTTATGTATATCCTTTACACCAACTTAGCTACTTGCTAAATTTTGATGTTAATGGTTATATTACGCAAGCAGCAGACTGTGGTTACAATGATTATACCCAGCTTGTCTGCTCGTATGGTGATTTTAATATGAAATCTGGTGTTTACTCTACTTCATATTATTCAGCCAAACCTGTAGGTGCGTACTATGAAGCTCATGTTTACCCAGATTGCAATTTTACTGATTTGTTCCGGGAAAATGCTCCCACAATCATGCAATATAAGCGTCAAGTTTTTACGCGTTGTAATTACAACCTCACACTACTGCTCTCTCTTGTGCAGGTGGATGAGTTTGTCTGCGATAAGATTACCCCTGAGGCTCTTGCAACAGGGTGTTATTCGTCTCTTACCGTCGATTGGTTTGCATTTCCGTATGCTTGGAAGTCATACCTAGCTATAGGTTCAGCAGATCGCATTGTGCGGTTTAATTATAACCAGGATTATAGCAATCCCTCTTGTAGAATTCACTCCAAGGTGAATTCTTCAGTTGGCATTTCTTACTCTGGTTTATATAGTTATATTACTAATTGTAATTACGGCGGCTTCAACAAGGATGACGTTGTTAAGCCTGGTGGTCGTGCCAGTCAACCCTGTGTTACTGGCGCACTCAATTCACCTACTAACGGTCAAGTCTGGTCTTTTAATTTTGGTGGCGTCCCTTACAGAACCTCCCGCCTCACCTACACTGACCATCTTAAAAACCCTCTAGATATGGTTTATGTCATCACTGTTAAGTATGAACCAGGCGCTGAAACTGTATGTCCCAAACAAGTGCGTCCTGATTATAGTACCAATATTACTGGCTTATTAGGCTCTTGTATCAGTTATGACATTTATGGTATAACTGGTACTGGTGTTTTCCAGCTGTGTAATGCAACTGGAATTCCTCAACAAAAGTTTGTCTATGACAAATTTGATAATATAATTGGCTTTCACTCTGATGACGGCAATTATTATTGTGTGGCACCTTGTGTCAGTGTGCCTGTTTCTGTTATATATGATGACAACACTAATCAATACGCCACATTGTTTGGCAGTGTTGCTTGTCAACATATATCTACAATGGCTGCTCAGTTTTCTCGTGAAACTCGTGCTTCCCTCGTTTCAAGAAATATGCAGAATCTTCTACAGACTTCTGTCGGTTGTGTCATGGGTTTCCATGAAACTAATGACACCGTTGAAGACTGCAATCTTTCTTTGGGACAATCACTCTGCGCAATCCCACCTAACACCAACTTGAGGGTTGGTCGCTCCACCTTTGGATTAGGTTCTTTAGCCTACAACAGTCCATTGCGTGTTGATGCACTTAACTCCTCTGAGTTTAAGGTCTCCTTGCCTCTCAATTTTACATTTGGTGTTACTCAGGAATATATTGAAACTAGCATACAGAAGATTACAGTTGACTGTAAACAGTACGTGTGCAATGGTTTTGCTAAGTGTGAAAAGCTGCTCGAACAATACGGTCAGTTTTGCTCTAAAATTAACCAGGCTCTCCATGGCGCGAATCTTCGCCAGGATGACTTTGTTCGTAATCTGTTTGAGAGTGTTAAAACACCACAGACAGTTCCTCTTACTACAGGTTTTGGAGGGGAGTTTAATCTTACTCTTCTAGAGCCGCTTTCTGTTTCTACAGGTTCTTCTAATGCGCGTAGTGCTTTGGAAGAGCTTTTGTTTGACAAAGTCACTATAGCTGATCCTGGCTACATGCAAGGTTATGATGACTGCATGCAACAGGGCCCTGCCTCAGCTCGTGATCTTATCTGTGCTCAATATGTTGCTGGCTACAAAGTGTTACCACCCCTTATGGATGTTAACATGGAAGCTGCATACACCTCTTCTTTGCTTGGTAGCATAGCTGGTGCTGGCTGGACTGCTGGTTTATCATCATTTGCTGCCATTCCATTTGCACAGAGTATATTTTACAGGTTAAATGGTGTTGGTATAACACAACAGGTTCTATCTGAGAATCAAAAGATTATTGCTAACAAGTTTAACCAAGCTCTTGGTGCCATGCAAACTGGTTTCACAACAACTAATGAAGCTTTTCAGAAAGTTCAAGATGCTGTGAACACTAATGCACAGGCTCTAGCTAAGTTGGCTAGTGAACTATCCAATACTTTTGGTGCTATTTCTTCTTCCATTGGTGACATCATTCAACGTCTTGATGTGCTTGAACAGGAAGTCCAAATAGACAGACTTATTAATGGCCGTCTGACTACACTCAACGCCTTTGTTGCTCAGCAGCTTGTTCGTTCTGAATCTGCTGCTCGTTCTGCACAATTGGCTAAGGATAAAGTCAATGAGTGTGTTAAATCACAATCCACTAGATCTGGATTTTGTGGTCAAGGCACTCATATAGTGTCCTTTGTTATTAACGCCCCTAATGGCCTCTACTTTATGCATGTTGGTTACCACCCTAGCCAACATATTGAGGTTGTTGCTGCCTATGGTCTTTGTGACGCGGCCAACCCCACTAATTGTATAGCCCCAGTTAATGGCTACTTTATTAAAAATCAAACTACTAGGGGTGTTGATGATTGGTCATATACAGGTTCTTCCTTTTATGCTCCAGAACCCATCACCACTCTTAATACTAGGTATGTCGCACCTCAAGTGACATTCCAAAACATTTCTACTAACCTTCCTCCTCCTCTGTTGGGCAATTCCACTGGAACTGACTTCAAAGATGAGTTGGATGAATTTTTCAAGAATGTTAGCACCAGTATACCAAATTTTGGTGCTCTAACACAAATTAATACTACTTTATTGGATCTTTCCGATGAAATGCTAGCTTTACAGCAAGTTGTCAAAGCGCTTAATGAGTCATATATTGACCTTAAAGAGCTCGGCAACTATACTTATTACAACAAATGGCCTTGGTACATTTGGTTGGGTTTCATTGCTGGACTTCTAGCCCTAGCCCTTTGTGTTTTCTTTATTCTTTGCTGCACTGGTTGCGGCACAAGTTGTTTAGGAAAACTTAAATGTAATCGTTGTTGTGACAAATATGAAGAATACGACCTTGAGCCGCATAAAATTCATATTCACTAA
Protein
MTYSVSLLMCLLTFIGANAKIVSIPGGVGTGACPQVDMQPSYFIKHNWPEPIDMNKADGVIYPNGRTYSNITLQTTNLFPRNGDLGTQYVYSASNEKSRTSNVAFISNYSYYGNPFGDGIVIRIGQNSNKTGSVIVGTAQTTIKKIYPALMLGSSFGNFSVNNKSGAYFNHTLLILPSKCGTVFQVAYCLLQPRTDSYCPGNANYVSYALIDSPTDCTSADESKRRNGLEDIKKYFNLVNCTYFEEFNVTADERAEWFGITQDSQGVHLYTSRKNGFNSNNLFLFASVPIYDKINYYTVIPRSIITPANQRSAWAAFYVYPLHQLSYLLNFDVNGYITQAADCGYNDYTQLVCSYGDFNMKSGVYSTSYYSAKPVGAYYEAHVYPDCNFTDLFRENAPTIMQYKRQVFTRCNYNLTLLLSLVQVDEFVCDKITPEALATGCYSSLTVDWFAFPYAWKSYLAIGSADRIVRFNYNQDYSNPSCRIHSKVNSSVGISYSGLYSYITNCNYGGFNKDDVVKPGGRASQPCVTGALNSPTNGQVWSFNFGGVPYRTSRLTYTDHLKNPLDMVYVITVKYEPGAETVCPKQVRPDYSTNITGLLGSCISYDIYGITGTGVFQLCNATGIPQQKFVYDKFDNIIGFHSDDGNYYCVAPCVSVPVSVIYDDNTNQYATLFGSVACQHISTMAAQFSRETRASLVSRNMQNLLQTSVGCVMGFHETNDTVEDCNLSLGQSLCAIPPNTNLRVGRSTFGLGSLAYNSPLRVDALNSSEFKVSLPLNFTFGVTQEYIETSIQKITVDCKQYVCNGFAKCEKLLEQYGQFCSKINQALHGANLRQDDFVRNLFESVKTPQTVPLTTGFGGEFNLTLLEPLSVSTGSSNARSALEELLFDKVTIADPGYMQGYDDCMQQGPASARDLICAQYVAGYKVLPPLMDVNMEAAYTSSLLGSIAGAGWTAGLSSFAAIPFAQSIFYRLNGVGITQQVLSENQKIIANKFNQALGAMQTGFTTTNEAFQKVQDAVNTNAQALAKLASELSNTFGAISSSIGDIIQRLDVLEQEVQIDRLINGRLTTLNAFVAQQLVRSESAARSAQLAKDKVNECVKSQSTRSGFCGQGTHIVSFVINAPNGLYFMHVGYHPSQHIEVVAAYGLCDAANPTNCIAPVNGYFIKNQTTRGVDDWSYTGSSFYAPEPITTLNTRYVAPQVTFQNISTNLPPPLLGNSTGTDFKDELDEFFKNVSTSIPNFGALTQINTTLLDLSDEMLALQQVVKALNESYIDLKELGNYTYYNKWPWYIWLGFIAGLLALALCVFFILCCTGCGTSCLGKLKCNRCCDKYEEYDLEPHKIHIH
Summary
Function
Spike protein S1: attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
attaches the virion to the cell membrane by interacting with host receptor, initiating the infection (By similarity). Interacts with host DPP4 to mediate virla entry.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
attaches the virion to the cell membrane by interacting with host receptor, initiating the infection (By similarity). Interacts with host DPP4 to mediate virla entry.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Subunit
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes.
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes. S1 interacts with murine DPP4.
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes. S1 interacts with murine DPP4.
Similarity
Belongs to the betacoronaviruses spike protein family.
Keywords
3D-structure
Coiled coil
Disulfide bond
Fusion of virus membrane with host endosomal membrane
Fusion of virus membrane with host membrane
Glycoprotein
Host cell membrane
Host membrane
Host-virus interaction
Lipoprotein
Membrane
Palmitate
Reference proteome
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Viral penetration into host cytoplasm
Virion
Virulence
Virus entry into host cell
Feature
chain Spike protein S2
Uniprot
A0A1W6ASU7
U5NJG5
A0A2R2YRG8
A0A2P1IT43
A0A2P1IT40
A0A140HF74
+ More
M4SVE7 A0A0U2P155 A0A0U2GL10 K0BRG7 A0A2D0YGK8 A0A384YDY2 A0A023VYK5 A0A2D0YNJ0 A0A482DX17 A0A1L2DZI5 A0A0U2MGJ8 A0A2D0YIQ0 A0A482DX85 A0A2P1IT75 A0A0S3IUB8 A0A1W5LIC4 A0A2H1YPP7 A0A2D0YDA3 A0A0U2NTX5 A0A4P8L6Q6 A0A0A0Q4U2 W6A0A7 A0A2D0Y380 A0A0U2NY03 A0A0U2NVR1 R9UCW7 A0A0U2P188 A0A291I6B7 A0A0H4JBS8 W5ZZF5 A0A384VXU1 W6A028 A0A411PBL8 A0A2D0YGA8 A0A0A0Q7F8 A0A2D0YAP4 A0A0U3SQE0 A0A0G3U597 A0A2D0YCE0 A0A0A0Q7G4 A0A0B5A1M3 A0A168DLI3 A0A411PBH5 A0A140AZ15 A0A140AYZ5 A0A0U2MS90 A0A410SR78 A0A140AZ05 A0A5C0PYB9 A0A2D0YPZ8 A0A516IIX0 A0A291I5Y3 A0A0U2J5U2 A0A0U2NY17 A0A0U4AGH8 T2B9T0 T2B999 A0A1W5LGN6 A0A3T0I8D8 A0A2H4GWK9 A0A0R9USA1 A0A411PBG2 A0A2D0Y9L1 A0A1W5LGL0 A0A1W5LGE4 M1RNL5 A0A0B5KTJ9 R9UQ53 T2BB24 A0A0U2GKS5 A0A291I632 A0A141BR51 K9N5Q8 A0A2H4KGX9 W6A8L9 T2BAV6 A0A411PBG4 A0A2P1IT84 A0A140AQ46 A0A0U2NE92 A0A2D0YLD5 A0A2D0YJ37 A0A024BSJ1 A0A3S7R2I2 A0A2D0YCT4 A0A2D0YM48 A0A126G7T9 A0A411PBD4 A0A0A0Q7F3 A0A3B8DUP5 A0A2D0YXA8 A0A2D0YUG7 A0A0U2P9D9 A0A2D0Y7Z5
M4SVE7 A0A0U2P155 A0A0U2GL10 K0BRG7 A0A2D0YGK8 A0A384YDY2 A0A023VYK5 A0A2D0YNJ0 A0A482DX17 A0A1L2DZI5 A0A0U2MGJ8 A0A2D0YIQ0 A0A482DX85 A0A2P1IT75 A0A0S3IUB8 A0A1W5LIC4 A0A2H1YPP7 A0A2D0YDA3 A0A0U2NTX5 A0A4P8L6Q6 A0A0A0Q4U2 W6A0A7 A0A2D0Y380 A0A0U2NY03 A0A0U2NVR1 R9UCW7 A0A0U2P188 A0A291I6B7 A0A0H4JBS8 W5ZZF5 A0A384VXU1 W6A028 A0A411PBL8 A0A2D0YGA8 A0A0A0Q7F8 A0A2D0YAP4 A0A0U3SQE0 A0A0G3U597 A0A2D0YCE0 A0A0A0Q7G4 A0A0B5A1M3 A0A168DLI3 A0A411PBH5 A0A140AZ15 A0A140AYZ5 A0A0U2MS90 A0A410SR78 A0A140AZ05 A0A5C0PYB9 A0A2D0YPZ8 A0A516IIX0 A0A291I5Y3 A0A0U2J5U2 A0A0U2NY17 A0A0U4AGH8 T2B9T0 T2B999 A0A1W5LGN6 A0A3T0I8D8 A0A2H4GWK9 A0A0R9USA1 A0A411PBG2 A0A2D0Y9L1 A0A1W5LGL0 A0A1W5LGE4 M1RNL5 A0A0B5KTJ9 R9UQ53 T2BB24 A0A0U2GKS5 A0A291I632 A0A141BR51 K9N5Q8 A0A2H4KGX9 W6A8L9 T2BAV6 A0A411PBG4 A0A2P1IT84 A0A140AQ46 A0A0U2NE92 A0A2D0YLD5 A0A2D0YJ37 A0A024BSJ1 A0A3S7R2I2 A0A2D0YCT4 A0A2D0YM48 A0A126G7T9 A0A411PBD4 A0A0A0Q7F3 A0A3B8DUP5 A0A2D0YXA8 A0A2D0YUG7 A0A0U2P9D9 A0A2D0Y7Z5
Pubmed
28377531
24050621
25031349
29579103
29507189
27125484
+ More
23903833 26678874 23170002 26391698 29116217 24874668 27999424 26632876 25323704 24549846 26281793 23782859 26079346 24857749 24896817 31553909 24781747 26584223 26272560 26691200 26981770 26839489 26933050 30753126 26272558 26814649 30482895 24055451 31022948 23693015 23835475 24473083 23078800 23486063 29239118 24786258 30294617
23903833 26678874 23170002 26391698 29116217 24874668 27999424 26632876 25323704 24549846 26281793 23782859 26079346 24857749 24896817 31553909 24781747 26584223 26272560 26691200 26981770 26839489 26933050 30753126 26272558 26814649 30482895 24055451 31022948 23693015 23835475 24473083 23078800 23486063 29239118 24786258 30294617
EMBL
KX574227
ARJ34226.1
KC869678
AGY29650.2
MF593268
ATQ39390.1
+ More
MG923466 MG923467 AVN89291.1 AVN89302.1 MG923468 AVN89313.1 KU740200 AMO03401.1 KC776174 AGH58717.1 KT805973 ALJ54453.1 KT368834 ALA49451.1 JX869059 AFS88936.1 MF598608 MF598650 MF598700 ASU90076.1 ASU90538.1 ASU91087.1 MG912607 AVK87450.1 KJ614529 AHY21469.1 MF598600 MF598678 ASU89988.1 ASU90846.1 MK564475 QBM11748.1 KX108943 ANI69889.1 KT806016 ALJ54496.1 MF598711 ASU91208.1 MK564474 QBM11737.1 MG923470 MG923471 AVN89334.1 AVN89344.1 KT805981 KT751244 MF598596 MF598599 MF598601 MF598605 MF598609 MF598610 MF598626 MF598630 MF598653 MF598655 MF598659 MF598671 MF598683 MF598690 MF598699 MF598702 MF598703 MF598704 MF598705 MF598706 MF598707 MF598708 MF598709 MF598710 MF598712 MF598713 MF598714 MF598715 MF598716 MF598717 MF598719 MF598720 MF598721 MF598722 ALJ54461.1 ALR69641.1 ASU89944.1 ASU89977.1 ASU89999.1 ASU90043.1 ASU90087.1 ASU90098.1 ASU90274.1 ASU90318.1 ASU90571.1 ASU90593.1 ASU90637.1 ASU90769.1 ASU90901.1 ASU90978.1 ASU91076.1 ASU91109.1 ASU91120.1 ASU91131.1 ASU91142.1 ASU91153.1 ASU91164.1 ASU91175.1 ASU91186.1 ASU91197.1 ASU91219.1 ASU91230.1 ASU91241.1 ASU91252.1 ASU91262.1 ASU91273.1 ASU91295.1 ASU91305.1 ASU91315.1 ASU91325.1 KX154689 MG757604 ANF29217.1 AXN73514.1 MF598597 ASU89955.1 MF598651 ASU90549.1 KT806028 ALJ54508.1 MK858160 MK858161 MK858162 QCQ28832.1 QCQ28833.1 QCQ28834.1 KM027279 KT368824 AID55090.1 ALA49341.1 KJ156905 AHI48692.1 MF598623 ASU90241.1 KT806020 ALJ54500.1 KT806011 ALJ54491.1 KF192507 AGN52936.1 KT806021 ALJ54501.1 MG011354 ATG84833.1 KP405225 KP405227 KT805988 MF598616 MG011341 MH822886 MK462250 AKO69634.1 AKO69636.1 ALJ54468.1 ASU90164.1 ATG84690.1 AYM48030.1 QBF80532.1 KF917527 KF958702 KJ156949 KJ156952 KJ556336 AHE78097.1 AHE78108.1 AHI48594.1 AHI48605.1 AHN10812.1 MF741827 MF741832 ASY99811.1 ASY99864.1 KJ156934 KJ156873 KJ713295 KJ713296 KM015348 KM027281 KM210278 KP966104 KT006149 KT026453 KT026454 KT026455 KT026456 KT036372 KT036373 KT036374 KT182953 KT275306 KT275307 KT275308 KT368833 KT368868 KT368870 KT368871 KT368872 KT368873 KT368874 KT368876 KT368877 KT368878 KT368882 KT368883 KT368884 KT368885 KT368886 KT368889 KT805964 KT805965 KT805967 KT805969 KT805974 KT805977 KT805979 KT805980 KT805982 KT805984 KT805989 KT805990 KT805993 KT805996 KT806002 KT806005 KT806007 KT806009 KT806012 KT806014 KT806018 KT806019 KT806023 KT806024 KT806025 KT806026 KT806027 KT806029 KT806030 KT806031 KT806033 KT806034 KT806035 KT806039 KT868870 KU233362 KU233363 KU233364 KT806044 KT806045 KT806046 KT806047 KT806048 KT806050 KT806051 KT806052 KT806053 KU851859 KU851860 KU851861 KU851862 KU851863 KU851864 KX154684 KX108945 KY688118 KY688120 KY688121 KY688122 KY688123 KY688124 MF598594 MF598595 MF598603 MF598604 MF598606 MF598607 MF598611 MF598612 MF598613 MF598615 MF598620 MF598625 MF598627 MF598628 MF598633 MF598643 MF598644 MF598645 MF598646 MF598652 MF598654 MF598657 MF598660 MF598661 MF598662 MF598664 MF598665 MF598666 MF598668 MF598670 MF598677 MF598680 MF598682 MF598684 MF598685 MF598686 MF598687 MF598688 MF598691 MF598692 MF598694 MF598695 MF598697 MF598698 MF598701 MF741837 MF741825 MF741826 MF741828 MF741829 MF741831 MF741833 MF741834 MF741835 MF741836 MG011343 MG011352 MG011353 MG011355 MF679172 MG366880 MG912608 MH306207 MH371127 MH432120 MG757593 MG757594 MG757595 MG757596 MG757598 MG757599 MG757600 MG757601 MG757602 MG757603 MH029552 MK129253 MH978888 MK462248 MH259486 MK858156 MK858158 MK858163 AHI48572.1 AHI48662.1 AHY22525.1 AHY22535.1 AID50418.1 AID55092.1 AJD81451.1 AKA63509.1 AKJ80137.2 AKK52582.1 AKK52592.1 AKK52602.1 AKK52612.1 AKL80593.1 AKL80604.1 AKL80615.1 AKN11071.1 AKQ21055.1 AKQ21064.1 AKQ21073.1 ALA49440.1 ALA49825.1 ALA49847.1 ALA49858.1 ALA49869.1 ALA49880.1 ALA49891.1 ALA49913.1 ALA49924.1 ALA49935.1 ALA49979.1 ALA49990.1 ALA50001.1 ALA50012.1 ALA50023.1 ALA50056.1 ALJ54444.1 ALJ54445.1 ALJ54447.1 ALJ54449.1 ALJ54454.1 ALJ54457.1 ALJ54459.1 ALJ54460.1 ALJ54462.1 ALJ54464.1 ALJ54469.1 ALJ54470.1 ALJ54473.1 ALJ54476.1 ALJ54482.1 ALJ54485.1 ALJ54487.1 ALJ54489.1 ALJ54492.1 ALJ54494.1 ALJ54498.1 ALJ54499.1 ALJ54503.1 ALJ54504.1 ALJ54505.1 ALJ54506.1 ALJ54507.1 ALJ54509.1 ALJ54510.1 ALJ54511.1 ALJ54513.1 ALJ54514.1 ALJ54515.1 ALJ54519.1 ALK80242.1 ALT66802.1 ALT66813.1 ALT66824.1 ALW82636.1 ALW82647.1 ALW82658.1 ALW82669.1 ALW82680.1 ALW82702.1 ALW82709.1 ALW82720.1 ALW82731.1 AMQ49015.1 AMQ49026.1 AMQ49037.1 AMQ49048.1 AMQ49059.1 AMQ49070.1 ANF29162.1 ANI69911.1 AQZ41311.1 AQZ41332.1 AQZ41343.1 AQZ41354.1 AQZ41365.1 AQZ41376.1 ASU89921.1 ASU89933.1 ASU90021.1 ASU90032.1 ASU90054.1 ASU90065.1 ASU90109.1 ASU90120.1 ASU90131.1 ASU90153.1 ASU90208.1 ASU90263.1 ASU90285.1 ASU90295.1 ASU90351.1 ASU90461.1 ASU90472.1 ASU90483.1 ASU90494.1 ASU90560.1 ASU90582.1 ASU90615.1 ASU90648.1 ASU90659.1 ASU90670.1 ASU90692.1 ASU90703.1 ASU90714.1 ASU90736.1 ASU90758.1 ASU90835.1 ASU90868.1 ASU90890.1 ASU90912.1 ASU90923.1 ASU90934.1 ASU90945.1 ASU90956.1 ASU90988.1 ASU90999.1 ASU91021.1 ASU91032.1 ASU91054.1 ASU91065.1 ASU91098.1 ASY99778.1 ASY99789.1 ASY99800.1 ASY99820.1 ASY99831.1 ASY99853.1 ASY99873.1 ASY99884.1 ASY99895.1 ASY99906.1 ATG84712.1 ATG84811.1 ATG84822.1 ATG84844.1 ATW75478.1 ATY74381.1 AVK87461.1 AWM99582.1 AXG21643.1 AXG21665.1 AXN73393.1 AXN73404.1 AXN73415.1 AXN73426.1 AXN73448.1 AXN73459.1 AXN73470.1 AXN73481.1 AXN73492.1 AXN73503.1 AYO86699.1 AZK15900.1 QBF44115.1 QBF80510.1 QCI31480.1 QCQ28828.1 QCQ28830.1 QCQ28835.1 MK462253 MK462254 MK462255 QBF80565.1 QBF80576.1 QBF80587.1 MF598629 MF598640 MF598658 MF598689 ASU90307.1 ASU90428.1 ASU90626.1 ASU90966.1 KM027273 AID55084.1 MF598679 ASU90857.1 KT806054 ALW82742.1 KT029139 AKL59401.1 MF598631 ASU90329.1 KM027288 AID55099.1 KM210277 AJD81440.1 KX034097 ANC28678.1 MK462247 QBF80499.1 KT868872 KT868876 ALK80261.1 ALK80301.1 KT182954 KT182955 KT326819 KT374052 KT374053 KT374054 KT374055 KT868865 KT868866 KT868867 KT868868 KT868869 KT868873 KT868874 KU308549 KX034096 KX034098 KX034099 KX034100 AKN11072.1 AKN11073.1 AKS48062.1 ALB08267.1 ALB08278.1 ALB08289.1 ALB08300.1 ALK80192.1 ALK80202.1 ALK80212.1 ALK80222.1 ALK80232.1 ALK80271.1 ALK80281.1 AML60270.1 ANC28667.1 ANC28689.1 ANC28700.1 ANC28711.1 KT805978 KT806003 ALJ54458.1 ALJ54483.1 MK357908 MK357909 QAT98898.1 QAT98909.1 KT868875 KX034094 KX034095 ALK80291.1 ANC28645.1 ANC28656.1 MN365232 MN365233 QEJ82215.1 QEJ82226.1 MF598632 ASU90340.1 MN120514 QDP16206.1 MG011342 ATG84701.1 KT806038 ALJ54518.1 KT806036 ALJ54516.1 KT806049 ALW82691.1 KF600652 KT806040 AGV08584.1 ALJ54520.1 KF600612 KF600620 AGV08379.1 AGV08408.1 KX154685 KX154688 MG011346 ANF29173.1 ANF29206.1 ATG84745.1 MK280984 AZU90731.1 KY673149 AQZ41296.1 KT225476 ALD51904.1 MK462251 QBF80543.1 MF598649 ASU90527.1 KX154691 ANF29239.1 KX154687 ANF29195.1 KC667074 AGG22542.1 KP719931 KP719932 AJG44102.1 AJG44113.1 KF186564 KF186566 KF186567 KF186565 KF600623 KF600627 KF600632 KF600636 KF600644 KF600645 KF600647 KF600651 KJ156866 KJ156872 KJ156939 AGN70929.1 AGN70951.1 AGN70962.1 AGN70973.1 AGV08426.1 AGV08444.1 AGV08480.1 AGV08505.1 AGV08535.1 AGV08546.1 AGV08558.1 AGV08573.1 AHI48517.1 AHI48652.1 AHI48711.1 KF600628 KJ156874 KJ156910 KJ156942 KJ156901 KP405226 AGV08455.1 AHI48539.1 AHI48561.1 AHI48626.1 AHI48682.1 AKO69635.1 KT368825 KT805983 ALA49352.1 ALJ54463.1 MG011348 MG011349 MG011350 ATG84767.1 ATG84778.1 ATG84789.1 KT182956 KT182957 AKN11074.1 AKN11075.1 KC164505 KY581694 ASU45807.1 KJ156881 AHI48550.1 KF600613 AGV08390.1 MK462246 QBF80488.1 MG923473 AVN89365.1 KT374056 ALB08311.1 KT806010 ALJ54490.1 MF598618 ASU90186.1 MF598634 ASU90362.1 KJ782549 AHZ20790.1 MG757597 AXN73437.1 MF598641 ASU90439.1 MF598638 ASU90406.1 KT374050 KT374051 ALB08246.1 ALB08257.1 MK462243 MK462244 MK462245 QBF80455.1 QBF80466.1 QBF80477.1 KM027255 KM027256 KM027257 KM027258 KM027259 KM027260 KM027261 KM027263 KM027264 KM027265 KM027266 KM027267 KM027269 KM027270 KM027271 KM027272 KM027275 KM027291 KT861629 KT861630 KT861631 KT861633 KT861628 KY581693 MK052676 MK039552 MK039553 AID55066.1 AID55067.1 AID55068.1 AID55069.1 AID55070.1 AID55071.1 AID55072.1 AID55074.1 AID55075.1 AID55076.1 AID55077.1 AID55078.1 AID55080.1 AID55081.1 AID55082.1 AID55083.1 AID55086.1 AID55102.1 ALX27225.1 ALX27226.1 ALX27227.1 ALX27229.1 ALX27243.1 ASU45796.1 AYP10298.1 AYV64549.1 AYV64560.1 MG520075 MG520076 MG546331 AXN73370.1 AYJ71481.1 MF598648 MF598667 MF598696 ASU90516.1 ASU90725.1 ASU91043.1 MF598635 MF598637 ASU90373.1 ASU90395.1 KT806008 KT806017 ALJ54488.1 ALJ54497.1 MF598619 MF598621 MF598624 MF598672 MF598675 ASU90197.1 ASU90219.1 ASU90252.1 ASU90780.1 ASU90813.1
MG923466 MG923467 AVN89291.1 AVN89302.1 MG923468 AVN89313.1 KU740200 AMO03401.1 KC776174 AGH58717.1 KT805973 ALJ54453.1 KT368834 ALA49451.1 JX869059 AFS88936.1 MF598608 MF598650 MF598700 ASU90076.1 ASU90538.1 ASU91087.1 MG912607 AVK87450.1 KJ614529 AHY21469.1 MF598600 MF598678 ASU89988.1 ASU90846.1 MK564475 QBM11748.1 KX108943 ANI69889.1 KT806016 ALJ54496.1 MF598711 ASU91208.1 MK564474 QBM11737.1 MG923470 MG923471 AVN89334.1 AVN89344.1 KT805981 KT751244 MF598596 MF598599 MF598601 MF598605 MF598609 MF598610 MF598626 MF598630 MF598653 MF598655 MF598659 MF598671 MF598683 MF598690 MF598699 MF598702 MF598703 MF598704 MF598705 MF598706 MF598707 MF598708 MF598709 MF598710 MF598712 MF598713 MF598714 MF598715 MF598716 MF598717 MF598719 MF598720 MF598721 MF598722 ALJ54461.1 ALR69641.1 ASU89944.1 ASU89977.1 ASU89999.1 ASU90043.1 ASU90087.1 ASU90098.1 ASU90274.1 ASU90318.1 ASU90571.1 ASU90593.1 ASU90637.1 ASU90769.1 ASU90901.1 ASU90978.1 ASU91076.1 ASU91109.1 ASU91120.1 ASU91131.1 ASU91142.1 ASU91153.1 ASU91164.1 ASU91175.1 ASU91186.1 ASU91197.1 ASU91219.1 ASU91230.1 ASU91241.1 ASU91252.1 ASU91262.1 ASU91273.1 ASU91295.1 ASU91305.1 ASU91315.1 ASU91325.1 KX154689 MG757604 ANF29217.1 AXN73514.1 MF598597 ASU89955.1 MF598651 ASU90549.1 KT806028 ALJ54508.1 MK858160 MK858161 MK858162 QCQ28832.1 QCQ28833.1 QCQ28834.1 KM027279 KT368824 AID55090.1 ALA49341.1 KJ156905 AHI48692.1 MF598623 ASU90241.1 KT806020 ALJ54500.1 KT806011 ALJ54491.1 KF192507 AGN52936.1 KT806021 ALJ54501.1 MG011354 ATG84833.1 KP405225 KP405227 KT805988 MF598616 MG011341 MH822886 MK462250 AKO69634.1 AKO69636.1 ALJ54468.1 ASU90164.1 ATG84690.1 AYM48030.1 QBF80532.1 KF917527 KF958702 KJ156949 KJ156952 KJ556336 AHE78097.1 AHE78108.1 AHI48594.1 AHI48605.1 AHN10812.1 MF741827 MF741832 ASY99811.1 ASY99864.1 KJ156934 KJ156873 KJ713295 KJ713296 KM015348 KM027281 KM210278 KP966104 KT006149 KT026453 KT026454 KT026455 KT026456 KT036372 KT036373 KT036374 KT182953 KT275306 KT275307 KT275308 KT368833 KT368868 KT368870 KT368871 KT368872 KT368873 KT368874 KT368876 KT368877 KT368878 KT368882 KT368883 KT368884 KT368885 KT368886 KT368889 KT805964 KT805965 KT805967 KT805969 KT805974 KT805977 KT805979 KT805980 KT805982 KT805984 KT805989 KT805990 KT805993 KT805996 KT806002 KT806005 KT806007 KT806009 KT806012 KT806014 KT806018 KT806019 KT806023 KT806024 KT806025 KT806026 KT806027 KT806029 KT806030 KT806031 KT806033 KT806034 KT806035 KT806039 KT868870 KU233362 KU233363 KU233364 KT806044 KT806045 KT806046 KT806047 KT806048 KT806050 KT806051 KT806052 KT806053 KU851859 KU851860 KU851861 KU851862 KU851863 KU851864 KX154684 KX108945 KY688118 KY688120 KY688121 KY688122 KY688123 KY688124 MF598594 MF598595 MF598603 MF598604 MF598606 MF598607 MF598611 MF598612 MF598613 MF598615 MF598620 MF598625 MF598627 MF598628 MF598633 MF598643 MF598644 MF598645 MF598646 MF598652 MF598654 MF598657 MF598660 MF598661 MF598662 MF598664 MF598665 MF598666 MF598668 MF598670 MF598677 MF598680 MF598682 MF598684 MF598685 MF598686 MF598687 MF598688 MF598691 MF598692 MF598694 MF598695 MF598697 MF598698 MF598701 MF741837 MF741825 MF741826 MF741828 MF741829 MF741831 MF741833 MF741834 MF741835 MF741836 MG011343 MG011352 MG011353 MG011355 MF679172 MG366880 MG912608 MH306207 MH371127 MH432120 MG757593 MG757594 MG757595 MG757596 MG757598 MG757599 MG757600 MG757601 MG757602 MG757603 MH029552 MK129253 MH978888 MK462248 MH259486 MK858156 MK858158 MK858163 AHI48572.1 AHI48662.1 AHY22525.1 AHY22535.1 AID50418.1 AID55092.1 AJD81451.1 AKA63509.1 AKJ80137.2 AKK52582.1 AKK52592.1 AKK52602.1 AKK52612.1 AKL80593.1 AKL80604.1 AKL80615.1 AKN11071.1 AKQ21055.1 AKQ21064.1 AKQ21073.1 ALA49440.1 ALA49825.1 ALA49847.1 ALA49858.1 ALA49869.1 ALA49880.1 ALA49891.1 ALA49913.1 ALA49924.1 ALA49935.1 ALA49979.1 ALA49990.1 ALA50001.1 ALA50012.1 ALA50023.1 ALA50056.1 ALJ54444.1 ALJ54445.1 ALJ54447.1 ALJ54449.1 ALJ54454.1 ALJ54457.1 ALJ54459.1 ALJ54460.1 ALJ54462.1 ALJ54464.1 ALJ54469.1 ALJ54470.1 ALJ54473.1 ALJ54476.1 ALJ54482.1 ALJ54485.1 ALJ54487.1 ALJ54489.1 ALJ54492.1 ALJ54494.1 ALJ54498.1 ALJ54499.1 ALJ54503.1 ALJ54504.1 ALJ54505.1 ALJ54506.1 ALJ54507.1 ALJ54509.1 ALJ54510.1 ALJ54511.1 ALJ54513.1 ALJ54514.1 ALJ54515.1 ALJ54519.1 ALK80242.1 ALT66802.1 ALT66813.1 ALT66824.1 ALW82636.1 ALW82647.1 ALW82658.1 ALW82669.1 ALW82680.1 ALW82702.1 ALW82709.1 ALW82720.1 ALW82731.1 AMQ49015.1 AMQ49026.1 AMQ49037.1 AMQ49048.1 AMQ49059.1 AMQ49070.1 ANF29162.1 ANI69911.1 AQZ41311.1 AQZ41332.1 AQZ41343.1 AQZ41354.1 AQZ41365.1 AQZ41376.1 ASU89921.1 ASU89933.1 ASU90021.1 ASU90032.1 ASU90054.1 ASU90065.1 ASU90109.1 ASU90120.1 ASU90131.1 ASU90153.1 ASU90208.1 ASU90263.1 ASU90285.1 ASU90295.1 ASU90351.1 ASU90461.1 ASU90472.1 ASU90483.1 ASU90494.1 ASU90560.1 ASU90582.1 ASU90615.1 ASU90648.1 ASU90659.1 ASU90670.1 ASU90692.1 ASU90703.1 ASU90714.1 ASU90736.1 ASU90758.1 ASU90835.1 ASU90868.1 ASU90890.1 ASU90912.1 ASU90923.1 ASU90934.1 ASU90945.1 ASU90956.1 ASU90988.1 ASU90999.1 ASU91021.1 ASU91032.1 ASU91054.1 ASU91065.1 ASU91098.1 ASY99778.1 ASY99789.1 ASY99800.1 ASY99820.1 ASY99831.1 ASY99853.1 ASY99873.1 ASY99884.1 ASY99895.1 ASY99906.1 ATG84712.1 ATG84811.1 ATG84822.1 ATG84844.1 ATW75478.1 ATY74381.1 AVK87461.1 AWM99582.1 AXG21643.1 AXG21665.1 AXN73393.1 AXN73404.1 AXN73415.1 AXN73426.1 AXN73448.1 AXN73459.1 AXN73470.1 AXN73481.1 AXN73492.1 AXN73503.1 AYO86699.1 AZK15900.1 QBF44115.1 QBF80510.1 QCI31480.1 QCQ28828.1 QCQ28830.1 QCQ28835.1 MK462253 MK462254 MK462255 QBF80565.1 QBF80576.1 QBF80587.1 MF598629 MF598640 MF598658 MF598689 ASU90307.1 ASU90428.1 ASU90626.1 ASU90966.1 KM027273 AID55084.1 MF598679 ASU90857.1 KT806054 ALW82742.1 KT029139 AKL59401.1 MF598631 ASU90329.1 KM027288 AID55099.1 KM210277 AJD81440.1 KX034097 ANC28678.1 MK462247 QBF80499.1 KT868872 KT868876 ALK80261.1 ALK80301.1 KT182954 KT182955 KT326819 KT374052 KT374053 KT374054 KT374055 KT868865 KT868866 KT868867 KT868868 KT868869 KT868873 KT868874 KU308549 KX034096 KX034098 KX034099 KX034100 AKN11072.1 AKN11073.1 AKS48062.1 ALB08267.1 ALB08278.1 ALB08289.1 ALB08300.1 ALK80192.1 ALK80202.1 ALK80212.1 ALK80222.1 ALK80232.1 ALK80271.1 ALK80281.1 AML60270.1 ANC28667.1 ANC28689.1 ANC28700.1 ANC28711.1 KT805978 KT806003 ALJ54458.1 ALJ54483.1 MK357908 MK357909 QAT98898.1 QAT98909.1 KT868875 KX034094 KX034095 ALK80291.1 ANC28645.1 ANC28656.1 MN365232 MN365233 QEJ82215.1 QEJ82226.1 MF598632 ASU90340.1 MN120514 QDP16206.1 MG011342 ATG84701.1 KT806038 ALJ54518.1 KT806036 ALJ54516.1 KT806049 ALW82691.1 KF600652 KT806040 AGV08584.1 ALJ54520.1 KF600612 KF600620 AGV08379.1 AGV08408.1 KX154685 KX154688 MG011346 ANF29173.1 ANF29206.1 ATG84745.1 MK280984 AZU90731.1 KY673149 AQZ41296.1 KT225476 ALD51904.1 MK462251 QBF80543.1 MF598649 ASU90527.1 KX154691 ANF29239.1 KX154687 ANF29195.1 KC667074 AGG22542.1 KP719931 KP719932 AJG44102.1 AJG44113.1 KF186564 KF186566 KF186567 KF186565 KF600623 KF600627 KF600632 KF600636 KF600644 KF600645 KF600647 KF600651 KJ156866 KJ156872 KJ156939 AGN70929.1 AGN70951.1 AGN70962.1 AGN70973.1 AGV08426.1 AGV08444.1 AGV08480.1 AGV08505.1 AGV08535.1 AGV08546.1 AGV08558.1 AGV08573.1 AHI48517.1 AHI48652.1 AHI48711.1 KF600628 KJ156874 KJ156910 KJ156942 KJ156901 KP405226 AGV08455.1 AHI48539.1 AHI48561.1 AHI48626.1 AHI48682.1 AKO69635.1 KT368825 KT805983 ALA49352.1 ALJ54463.1 MG011348 MG011349 MG011350 ATG84767.1 ATG84778.1 ATG84789.1 KT182956 KT182957 AKN11074.1 AKN11075.1 KC164505 KY581694 ASU45807.1 KJ156881 AHI48550.1 KF600613 AGV08390.1 MK462246 QBF80488.1 MG923473 AVN89365.1 KT374056 ALB08311.1 KT806010 ALJ54490.1 MF598618 ASU90186.1 MF598634 ASU90362.1 KJ782549 AHZ20790.1 MG757597 AXN73437.1 MF598641 ASU90439.1 MF598638 ASU90406.1 KT374050 KT374051 ALB08246.1 ALB08257.1 MK462243 MK462244 MK462245 QBF80455.1 QBF80466.1 QBF80477.1 KM027255 KM027256 KM027257 KM027258 KM027259 KM027260 KM027261 KM027263 KM027264 KM027265 KM027266 KM027267 KM027269 KM027270 KM027271 KM027272 KM027275 KM027291 KT861629 KT861630 KT861631 KT861633 KT861628 KY581693 MK052676 MK039552 MK039553 AID55066.1 AID55067.1 AID55068.1 AID55069.1 AID55070.1 AID55071.1 AID55072.1 AID55074.1 AID55075.1 AID55076.1 AID55077.1 AID55078.1 AID55080.1 AID55081.1 AID55082.1 AID55083.1 AID55086.1 AID55102.1 ALX27225.1 ALX27226.1 ALX27227.1 ALX27229.1 ALX27243.1 ASU45796.1 AYP10298.1 AYV64549.1 AYV64560.1 MG520075 MG520076 MG546331 AXN73370.1 AYJ71481.1 MF598648 MF598667 MF598696 ASU90516.1 ASU90725.1 ASU91043.1 MF598635 MF598637 ASU90373.1 ASU90395.1 KT806008 KT806017 ALJ54488.1 ALJ54497.1 MF598619 MF598621 MF598624 MF598672 MF598675 ASU90197.1 ASU90219.1 ASU90252.1 ASU90780.1 ASU90813.1
Proteomes
UP000202992
UP000145931
UP000108469
UP000168352
UP000112279
UP000180345
+ More
UP000135646 UP000138672 UP000104220 UP000119765 UP000120438 UP000135014 UP000148657 UP000159930 UP000095864 UP000096875 UP000102966 UP000104540 UP000109666 UP000112022 UP000115322 UP000118563 UP000119223 UP000121965 UP000122049 UP000125198 UP000128184 UP000128186 UP000128651 UP000142239 UP000144056 UP000144472 UP000147538 UP000151795 UP000153269 UP000157492 UP000158935 UP000159306 UP000161565 UP000163035 UP000163895 UP000170921 UP000171516 UP000171892 UP000174054 UP000174558 UP000180347 UP000180348 UP000171389 UP000118772 UP000114773 UP000129370 UP000097149 UP000098933 UP000105311 UP000106595 UP000109122 UP000126763 UP000128076 UP000140611 UP000147342 UP000145271 UP000167218 UP000146591 UP000148810 UP000140430 UP000154518 UP000119183 UP000160415 UP000131416 UP000174176 UP000101567 UP000108647 UP000117251 UP000125629 UP000127508 UP000130711 UP000140498 UP000144449 UP000158466 UP000160534 UP000171868 UP000115754 UP000165701 UP000167295 UP000131256 UP000139997 UP000105272 UP000134122 UP000149108 UP000103178 UP000113724 UP000100765 UP000113762 UP000116785 UP000124216 UP000144709 UP000159190 UP000175153
UP000135646 UP000138672 UP000104220 UP000119765 UP000120438 UP000135014 UP000148657 UP000159930 UP000095864 UP000096875 UP000102966 UP000104540 UP000109666 UP000112022 UP000115322 UP000118563 UP000119223 UP000121965 UP000122049 UP000125198 UP000128184 UP000128186 UP000128651 UP000142239 UP000144056 UP000144472 UP000147538 UP000151795 UP000153269 UP000157492 UP000158935 UP000159306 UP000161565 UP000163035 UP000163895 UP000170921 UP000171516 UP000171892 UP000174054 UP000174558 UP000180347 UP000180348 UP000171389 UP000118772 UP000114773 UP000129370 UP000097149 UP000098933 UP000105311 UP000106595 UP000109122 UP000126763 UP000128076 UP000140611 UP000147342 UP000145271 UP000167218 UP000146591 UP000148810 UP000140430 UP000154518 UP000119183 UP000160415 UP000131416 UP000174176 UP000101567 UP000108647 UP000117251 UP000125629 UP000127508 UP000130711 UP000140498 UP000144449 UP000158466 UP000160534 UP000171868 UP000115754 UP000165701 UP000167295 UP000131256 UP000139997 UP000105272 UP000134122 UP000149108 UP000103178 UP000113724 UP000100765 UP000113762 UP000116785 UP000124216 UP000144709 UP000159190 UP000175153
PRIDE
Interpro
SUPFAM
SSF143587
SSF143587
Gene 3D
ProteinModelPortal
A0A1W6ASU7
U5NJG5
A0A2R2YRG8
A0A2P1IT43
A0A2P1IT40
A0A140HF74
+ More
M4SVE7 A0A0U2P155 A0A0U2GL10 K0BRG7 A0A2D0YGK8 A0A384YDY2 A0A023VYK5 A0A2D0YNJ0 A0A482DX17 A0A1L2DZI5 A0A0U2MGJ8 A0A2D0YIQ0 A0A482DX85 A0A2P1IT75 A0A0S3IUB8 A0A1W5LIC4 A0A2H1YPP7 A0A2D0YDA3 A0A0U2NTX5 A0A4P8L6Q6 A0A0A0Q4U2 W6A0A7 A0A2D0Y380 A0A0U2NY03 A0A0U2NVR1 R9UCW7 A0A0U2P188 A0A291I6B7 A0A0H4JBS8 W5ZZF5 A0A384VXU1 W6A028 A0A411PBL8 A0A2D0YGA8 A0A0A0Q7F8 A0A2D0YAP4 A0A0U3SQE0 A0A0G3U597 A0A2D0YCE0 A0A0A0Q7G4 A0A0B5A1M3 A0A168DLI3 A0A411PBH5 A0A140AZ15 A0A140AYZ5 A0A0U2MS90 A0A410SR78 A0A140AZ05 A0A5C0PYB9 A0A2D0YPZ8 A0A516IIX0 A0A291I5Y3 A0A0U2J5U2 A0A0U2NY17 A0A0U4AGH8 T2B9T0 T2B999 A0A1W5LGN6 A0A3T0I8D8 A0A2H4GWK9 A0A0R9USA1 A0A411PBG2 A0A2D0Y9L1 A0A1W5LGL0 A0A1W5LGE4 M1RNL5 A0A0B5KTJ9 R9UQ53 T2BB24 A0A0U2GKS5 A0A291I632 A0A141BR51 K9N5Q8 A0A2H4KGX9 W6A8L9 T2BAV6 A0A411PBG4 A0A2P1IT84 A0A140AQ46 A0A0U2NE92 A0A2D0YLD5 A0A2D0YJ37 A0A024BSJ1 A0A3S7R2I2 A0A2D0YCT4 A0A2D0YM48 A0A126G7T9 A0A411PBD4 A0A0A0Q7F3 A0A3B8DUP5 A0A2D0YXA8 A0A2D0YUG7 A0A0U2P9D9 A0A2D0Y7Z5
M4SVE7 A0A0U2P155 A0A0U2GL10 K0BRG7 A0A2D0YGK8 A0A384YDY2 A0A023VYK5 A0A2D0YNJ0 A0A482DX17 A0A1L2DZI5 A0A0U2MGJ8 A0A2D0YIQ0 A0A482DX85 A0A2P1IT75 A0A0S3IUB8 A0A1W5LIC4 A0A2H1YPP7 A0A2D0YDA3 A0A0U2NTX5 A0A4P8L6Q6 A0A0A0Q4U2 W6A0A7 A0A2D0Y380 A0A0U2NY03 A0A0U2NVR1 R9UCW7 A0A0U2P188 A0A291I6B7 A0A0H4JBS8 W5ZZF5 A0A384VXU1 W6A028 A0A411PBL8 A0A2D0YGA8 A0A0A0Q7F8 A0A2D0YAP4 A0A0U3SQE0 A0A0G3U597 A0A2D0YCE0 A0A0A0Q7G4 A0A0B5A1M3 A0A168DLI3 A0A411PBH5 A0A140AZ15 A0A140AYZ5 A0A0U2MS90 A0A410SR78 A0A140AZ05 A0A5C0PYB9 A0A2D0YPZ8 A0A516IIX0 A0A291I5Y3 A0A0U2J5U2 A0A0U2NY17 A0A0U4AGH8 T2B9T0 T2B999 A0A1W5LGN6 A0A3T0I8D8 A0A2H4GWK9 A0A0R9USA1 A0A411PBG2 A0A2D0Y9L1 A0A1W5LGL0 A0A1W5LGE4 M1RNL5 A0A0B5KTJ9 R9UQ53 T2BB24 A0A0U2GKS5 A0A291I632 A0A141BR51 K9N5Q8 A0A2H4KGX9 W6A8L9 T2BAV6 A0A411PBG4 A0A2P1IT84 A0A140AQ46 A0A0U2NE92 A0A2D0YLD5 A0A2D0YJ37 A0A024BSJ1 A0A3S7R2I2 A0A2D0YCT4 A0A2D0YM48 A0A126G7T9 A0A411PBD4 A0A0A0Q7F3 A0A3B8DUP5 A0A2D0YXA8 A0A2D0YUG7 A0A0U2P9D9 A0A2D0Y7Z5
PDB
5X5F
E-value=0,
Score=4044
Ontologies
GO
GO:0061025 P:membrane fusion
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0016021 C:integral component of membrane
GO:0019031 C:viral envelope
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0020002 C:host cell plasma membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0016021 C:integral component of membrane
GO:0019031 C:viral envelope
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0020002 C:host cell plasma membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
Subcellular Location
From MSLVP
Host nucleus > host nucleolus. Host cytoplasm. Secreted.
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
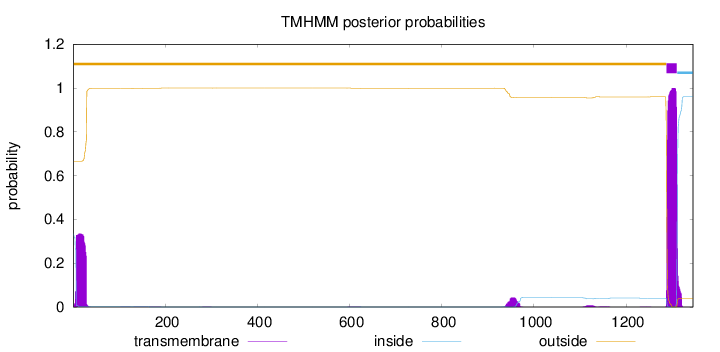
Topology
Length:
1345
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
32.26825
Exp number, first 60 AAs:
7.54774
Total prob of N-in:
0.33524
outside
1 - 1287
TMhelix
1288 - 1310
inside
1311 - 1345
Population Genetic Test Statistics
Pi
0.385847
Theta
0.4241027
Tajima's D
-0.5008172
CLR
74.27303
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 |
Protein |
YP_009361857.1 |
|
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.