Strain
CHINA_MURINE_2011_NC_034972
(Region: China; Strain: Coronavirus AcCoV-JC34, complete genome.; Date: 2011)
Gene
membrane protein
Description
Annotated in NCBI,
membrane protein
Location
GenBank Accession
Full name
Membrane protein
Alternative Name
E1 glycoprotein
Matrix glycoprotein
Membrane glycoprotein
Matrix glycoprotein
Membrane glycoprotein
Sequence
CDS
ATGGTACTCTTTTGTGAGTTTAATGACACTGCCACCAATGGCTGTGAAGCCTGTGTTAATCAGGGCTGCACAGGTAATGGTGTCACTAGTGCCAGCATCACTTGGCATTTGCTGAACTGGAACTTTAGCTGGAGTGTTATACTCACTGTTTTTGTTGCAGTTCTGCAGTATGGCAATCTTAAGTACAGCATGATTCTGTACATCCTTAAAATGATCATCATGTGGTTGCTATGGCCTATTGTCATAGCACTAACCATTTTTGATGCCATTGAAACCTTCAATCAGGAGCGCTATGTCATGTTTGGATTCAGCATTGCTCTTGGTGTGGTCACGTTGATACTCTGGATTATGTACTTCGTAAACAGCTTTAGGCTTTACAGACGTACTAACTCCTGGTGGAGTTTCAACCCAGAGACCAATGCCATAATCTGTGTTTCAGTCATGGGGCGAAACATAACCATGCCAACCCCAGTCTGCCCCACAGGTGTCACACTAACGCTTCTTTCAGGAACGCTCTATGTTGAAGGGCAACGTGCTGCCACAGGCGTGAATCTTGACAATCTCCCTAGTTACGTTACTGTAGCAAAACCTGGAGTCACTATAGTTTACCACCGTGTTGGTAAATCAGTACGTGTCTCCACAGCTACGGGATGGATGTACTATGTCAAGAGTAAAGCAGGTGATTATTCTGCCTACTCCAGTGGTGGCAATCTATCAGATCAGGAACAACTGCTTCACATGGTCTAA
Protein
MVLFCEFNDTATNGCEACVNQGCTGNGVTSASITWHLLNWNFSWSVILTVFVAVLQYGNLKYSMILYILKMIIMWLLWPIVIALTIFDAIETFNQERYVMFGFSIALGVVTLILWIMYFVNSFRLYRRTNSWWSFNPETNAIICVSVMGRNITMPTPVCPTGVTLTLLSGTLYVEGQRAATGVNLDNLPSYVTVAKPGVTIVYHRVGKSVRVSTATGWMYYVKSKAGDYSAYSSGGNLSDQEQLLHMV
Summary
Function
Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins.
Subunit
Homomultimer. Interacts with envelope E protein in the budding compartment of the host cell, which is located between endoplasmic reticulum and the Golgi complex. Forms a complex with HE and S proteins. Interacts with nucleocapsid N protein. This interaction probably participates in RNA packaging into the virus.
Similarity
Belongs to the alphacoronaviruses M protein family.
Keywords
Glycoprotein
Host Golgi apparatus
Host membrane
Membrane
Transmembrane
Transmembrane helix
Viral envelope protein
Viral matrix protein
Virion
Reference proteome
Feature
chain Membrane protein
Uniprot
A0A1X9JJV5
A0A2H4MX25
A0A2H4MXE5
A0A1L7HJ65
A0A2H4MWY4
A0A481S0S6
+ More
G8J7G9 F6M1R4 B7T076 B7T066 Q5FX57 Q6TPL0 A0A1D8ELQ6 Q64FR0 B7T4B3 A0A0U2TVR6 Q9DY23 G8A505 M4I7F1 A0A384VGP5 A0A222UC68 H9TEW2 Q6TPL1 A0A0D5ZYB2 M4I7F5 A0A0Y0R0E2 Q0PL26 Q5FX58 Q0PKZ1 A0A384V9R1 A0A384VA81 A0A384V9Q6 R9RZN0 Q88514 A7UJ02 A0A384VIC0 Q7T713 A0A384V9L3 M4I824 Q6PQ04 C0J8Y5 P33464 A0A384V9G9 A0A384V9E4 A0A3S6JCJ2 Q9WKM6 E7E031 Q0PL00 F1CNZ1 P04135 Q5G1S6 A0A514KFM3 A0A514KFK1 S6B903 Q1W2N8 A0A1S6KDW8 D2WXM2 S6CE17 A0A1L7NS55 Q5D578 Q0ZVX1 H9TEX6 Q5G1S5 G3CI08 A0A172B6J4 I3QKR3 U3Q0B8 A0A1B4X968 Q0PL09 C6K2U0 A0A514KFL5 A0A224ANT6 A0A0K2SF30 A0A0E3GM58 Q5DQ89 A0A0K2BMP6 A0A514KFJ6 A0A1L6BQB0 P24412 D3XGJ7 A0A222UBY5 Q09PH1 A0A514KHE6 A0A514KH05 A0A514KFK0 A0A514KFM0 P09175 A0A514KFK2 Q6T524 Q64FQ9 Q7T6S9 Q19NU4 A0A088ND96 Q80RP1 C6GHD0 Q8JVR1 C5IJ46 E0AH02 C5IJ38
G8J7G9 F6M1R4 B7T076 B7T066 Q5FX57 Q6TPL0 A0A1D8ELQ6 Q64FR0 B7T4B3 A0A0U2TVR6 Q9DY23 G8A505 M4I7F1 A0A384VGP5 A0A222UC68 H9TEW2 Q6TPL1 A0A0D5ZYB2 M4I7F5 A0A0Y0R0E2 Q0PL26 Q5FX58 Q0PKZ1 A0A384V9R1 A0A384VA81 A0A384V9Q6 R9RZN0 Q88514 A7UJ02 A0A384VIC0 Q7T713 A0A384V9L3 M4I824 Q6PQ04 C0J8Y5 P33464 A0A384V9G9 A0A384V9E4 A0A3S6JCJ2 Q9WKM6 E7E031 Q0PL00 F1CNZ1 P04135 Q5G1S6 A0A514KFM3 A0A514KFK1 S6B903 Q1W2N8 A0A1S6KDW8 D2WXM2 S6CE17 A0A1L7NS55 Q5D578 Q0ZVX1 H9TEX6 Q5G1S5 G3CI08 A0A172B6J4 I3QKR3 U3Q0B8 A0A1B4X968 Q0PL09 C6K2U0 A0A514KFL5 A0A224ANT6 A0A0K2SF30 A0A0E3GM58 Q5DQ89 A0A0K2BMP6 A0A514KFJ6 A0A1L6BQB0 P24412 D3XGJ7 A0A222UBY5 Q09PH1 A0A514KHE6 A0A514KH05 A0A514KFK0 A0A514KFM0 P09175 A0A514KFK2 Q6T524 Q64FQ9 Q7T6S9 Q19NU4 A0A088ND96 Q80RP1 C6GHD0 Q8JVR1 C5IJ46 E0AH02 C5IJ38
Pubmed
28549438
30285857
25463600
30704076
22609354
22002680
+ More
19036814 18306032 27619563 21481551 22542271 25953186 17023013 25890036 8546012 20229183 19162099 1662846 10365166 2841792 3035066 2825819 10805807 3008432 11152504 1309909 7636969 25180686 27840394 16704791 17275120 23043168 20603153 26221765 2174956 20079778 2845226 15177889 12505636 8839428
19036814 18306032 27619563 21481551 22542271 25953186 17023013 25890036 8546012 20229183 19162099 1662846 10365166 2841792 3035066 2825819 10805807 3008432 11152504 1309909 7636969 25180686 27840394 16704791 17275120 23043168 20603153 26221765 2174956 20079778 2845226 15177889 12505636 8839428
EMBL
KX964649
AQW43026.1
KY370054
ATP66791.1
KY370050
ATP66769.1
+ More
KF294380 APU57647.1 KY370045 ATP66740.1 MK163627 QBG64660.1 JN856008 AEQ61972.1 JF682842 AEE69327.1 EU856362 ACJ63247.1 EU856361 ACJ63236.1 AY884049 KC175340 KC175341 AAW57899.1 AFX81102.1 AFX81112.1 AY390344 AAQ96368.1 KX689261 AOT21923.1 AY704916 AAU29609.1 EU924790 EU924791 HQ450377 ACJ64180.1 ACJ64189.1 ADU17739.1 KR061459 ALR84918.1 AF302262 AAG30226.1 GU146061 ADB10854.1 JQ693056 AFZ88851.1 KX900410 ASV64433.1 KX512809 ASR18941.1 JQ404409 AFG19730.1 AY390343 AAQ96367.1 KP981644 AKA65834.1 JQ693055 AFZ88850.1 KT696544 AMB66492.1 DQ811785 ABG89300.1 AY884048 AAW57898.1 DQ811789 ABG89329.1 KX900408 KX900409 ASV64415.1 ASV64424.1 KX900397 ASV64317.1 KX900396 KX900399 KX900400 KX900401 KX900403 KX900405 KX900406 KX900407 ASV64308.1 ASV64335.1 ASV64343.1 ASV64353.1 ASV64371.1 ASV64388.1 ASV64397.1 ASV64406.1 KC962433 AGM75130.1 Z35758 CAA84810.1 EU074218 FJ755618 ABU49663.1 ACN71200.1 KX900411 ASV64442.1 AY335549 DQ201447 AAQ02626.1 ABC72418.1 KX900404 ASV64379.1 JQ693053 AFZ88848.1 AY587883 AAT00646.1 FJ009114 ACN60154.1 X55980 X60056 KX900398 ASV64326.1 KX900395 ASV64299.1 KY406735 ASV45152.1 AF104420 AAC96007.1 HQ450376 ADU17730.1 DQ811788 ABG89323.1 HQ462571 ADY39744.1 M21627 X05598 X06371 AJ271965 M14878 AY864661 AAW66660.1 MK986745 QDI78404.1 MK986743 QDI78402.1 AB781790 BAN67923.1 DQ443743 ABD97839.1 JQ693054 KU729220 KX499468 KX900393 KX900394 KX900402 MK166015 AFZ88849.1 AML22781.1 AQT01353.1 ASV64281.1 ASV64290.1 ASV64362.1 QCZ41128.1 GQ477367 ADB28912.1 AB781788 BAN67905.1 LC190907 BAW32707.1 AY899209 AAX18705.1 DQ112226 AAZ91441.1 JQ404410 KY063616 KY063617 KY063618 AFG19744.1 ASB15743.1 ASB15753.1 ASB15761.1 AY864662 AAW66661.1 HM776941 AEM55570.1 KM347965 AKG92643.1 JQ700303 AFK10485.1 KF273109 AGW80452.1 LC119077 BAV31352.1 DQ811787 ABG89312.1 GQ152141 ACS44222.1 MK986744 QDI78403.1 LC215871 BBA21171.1 LC029419 BAS25713.1 KP202848 AKA60058.1 AY548235 AAT45197.1 KP849472 AKZ66477.1 MK986737 QDI78396.1 KX083668 APQ31246.1 Z24675 GU338457 ADD49361.1 KX512810 ASR18949.1 DQ848678 ABI14450.1 MK986742 QDI78401.1 MK986746 QDI78405.1 MK986739 QDI78398.1 MK986741 QDI78400.1 Y00560 MK986740 QDI78399.1 AY436635 AAR11075.1 AY704917 AAU29610.1 AY342160 DQ517438 ABF72147.1 KJ879395 AIN55916.1 AF502583 AAO33711.1 FJ938055 ACT10900.1 AB086903 BAC01158.1 FJ943762 ACR46122.1 HQ012372 ADL71502.1 FJ943761 ACR46114.1
KF294380 APU57647.1 KY370045 ATP66740.1 MK163627 QBG64660.1 JN856008 AEQ61972.1 JF682842 AEE69327.1 EU856362 ACJ63247.1 EU856361 ACJ63236.1 AY884049 KC175340 KC175341 AAW57899.1 AFX81102.1 AFX81112.1 AY390344 AAQ96368.1 KX689261 AOT21923.1 AY704916 AAU29609.1 EU924790 EU924791 HQ450377 ACJ64180.1 ACJ64189.1 ADU17739.1 KR061459 ALR84918.1 AF302262 AAG30226.1 GU146061 ADB10854.1 JQ693056 AFZ88851.1 KX900410 ASV64433.1 KX512809 ASR18941.1 JQ404409 AFG19730.1 AY390343 AAQ96367.1 KP981644 AKA65834.1 JQ693055 AFZ88850.1 KT696544 AMB66492.1 DQ811785 ABG89300.1 AY884048 AAW57898.1 DQ811789 ABG89329.1 KX900408 KX900409 ASV64415.1 ASV64424.1 KX900397 ASV64317.1 KX900396 KX900399 KX900400 KX900401 KX900403 KX900405 KX900406 KX900407 ASV64308.1 ASV64335.1 ASV64343.1 ASV64353.1 ASV64371.1 ASV64388.1 ASV64397.1 ASV64406.1 KC962433 AGM75130.1 Z35758 CAA84810.1 EU074218 FJ755618 ABU49663.1 ACN71200.1 KX900411 ASV64442.1 AY335549 DQ201447 AAQ02626.1 ABC72418.1 KX900404 ASV64379.1 JQ693053 AFZ88848.1 AY587883 AAT00646.1 FJ009114 ACN60154.1 X55980 X60056 KX900398 ASV64326.1 KX900395 ASV64299.1 KY406735 ASV45152.1 AF104420 AAC96007.1 HQ450376 ADU17730.1 DQ811788 ABG89323.1 HQ462571 ADY39744.1 M21627 X05598 X06371 AJ271965 M14878 AY864661 AAW66660.1 MK986745 QDI78404.1 MK986743 QDI78402.1 AB781790 BAN67923.1 DQ443743 ABD97839.1 JQ693054 KU729220 KX499468 KX900393 KX900394 KX900402 MK166015 AFZ88849.1 AML22781.1 AQT01353.1 ASV64281.1 ASV64290.1 ASV64362.1 QCZ41128.1 GQ477367 ADB28912.1 AB781788 BAN67905.1 LC190907 BAW32707.1 AY899209 AAX18705.1 DQ112226 AAZ91441.1 JQ404410 KY063616 KY063617 KY063618 AFG19744.1 ASB15743.1 ASB15753.1 ASB15761.1 AY864662 AAW66661.1 HM776941 AEM55570.1 KM347965 AKG92643.1 JQ700303 AFK10485.1 KF273109 AGW80452.1 LC119077 BAV31352.1 DQ811787 ABG89312.1 GQ152141 ACS44222.1 MK986744 QDI78403.1 LC215871 BBA21171.1 LC029419 BAS25713.1 KP202848 AKA60058.1 AY548235 AAT45197.1 KP849472 AKZ66477.1 MK986737 QDI78396.1 KX083668 APQ31246.1 Z24675 GU338457 ADD49361.1 KX512810 ASR18949.1 DQ848678 ABI14450.1 MK986742 QDI78401.1 MK986746 QDI78405.1 MK986739 QDI78398.1 MK986741 QDI78400.1 Y00560 MK986740 QDI78399.1 AY436635 AAR11075.1 AY704917 AAU29610.1 AY342160 DQ517438 ABF72147.1 KJ879395 AIN55916.1 AF502583 AAO33711.1 FJ938055 ACT10900.1 AB086903 BAC01158.1 FJ943762 ACR46122.1 HQ012372 ADL71502.1 FJ943761 ACR46114.1
Proteomes
UP000203395
UP000204448
UP000153699
UP000155481
UP000165378
UP000125818
+ More
UP000119169 UP000115330 UP000166023 UP000145148 UP000158285 UP000123026 UP000111647 UP000121388 UP000156732 UP000137024 UP000113525 UP000001440 UP000158504 UP000131894 UP000129066 UP000106470 UP000166047 UP000150460 UP000272061 UP000149523 UP000164750 UP000098772 UP000174600 UP000118215 UP000162682 UP000110620
UP000119169 UP000115330 UP000166023 UP000145148 UP000158285 UP000123026 UP000111647 UP000121388 UP000156732 UP000137024 UP000113525 UP000001440 UP000158504 UP000131894 UP000129066 UP000106470 UP000166047 UP000150460 UP000272061 UP000149523 UP000164750 UP000098772 UP000174600 UP000118215 UP000162682 UP000110620
Pfam
PF01635
Corona_M
ProteinModelPortal
A0A1X9JJV5
A0A2H4MX25
A0A2H4MXE5
A0A1L7HJ65
A0A2H4MWY4
A0A481S0S6
+ More
G8J7G9 F6M1R4 B7T076 B7T066 Q5FX57 Q6TPL0 A0A1D8ELQ6 Q64FR0 B7T4B3 A0A0U2TVR6 Q9DY23 G8A505 M4I7F1 A0A384VGP5 A0A222UC68 H9TEW2 Q6TPL1 A0A0D5ZYB2 M4I7F5 A0A0Y0R0E2 Q0PL26 Q5FX58 Q0PKZ1 A0A384V9R1 A0A384VA81 A0A384V9Q6 R9RZN0 Q88514 A7UJ02 A0A384VIC0 Q7T713 A0A384V9L3 M4I824 Q6PQ04 C0J8Y5 P33464 A0A384V9G9 A0A384V9E4 A0A3S6JCJ2 Q9WKM6 E7E031 Q0PL00 F1CNZ1 P04135 Q5G1S6 A0A514KFM3 A0A514KFK1 S6B903 Q1W2N8 A0A1S6KDW8 D2WXM2 S6CE17 A0A1L7NS55 Q5D578 Q0ZVX1 H9TEX6 Q5G1S5 G3CI08 A0A172B6J4 I3QKR3 U3Q0B8 A0A1B4X968 Q0PL09 C6K2U0 A0A514KFL5 A0A224ANT6 A0A0K2SF30 A0A0E3GM58 Q5DQ89 A0A0K2BMP6 A0A514KFJ6 A0A1L6BQB0 P24412 D3XGJ7 A0A222UBY5 Q09PH1 A0A514KHE6 A0A514KH05 A0A514KFK0 A0A514KFM0 P09175 A0A514KFK2 Q6T524 Q64FQ9 Q7T6S9 Q19NU4 A0A088ND96 Q80RP1 C6GHD0 Q8JVR1 C5IJ46 E0AH02 C5IJ38
G8J7G9 F6M1R4 B7T076 B7T066 Q5FX57 Q6TPL0 A0A1D8ELQ6 Q64FR0 B7T4B3 A0A0U2TVR6 Q9DY23 G8A505 M4I7F1 A0A384VGP5 A0A222UC68 H9TEW2 Q6TPL1 A0A0D5ZYB2 M4I7F5 A0A0Y0R0E2 Q0PL26 Q5FX58 Q0PKZ1 A0A384V9R1 A0A384VA81 A0A384V9Q6 R9RZN0 Q88514 A7UJ02 A0A384VIC0 Q7T713 A0A384V9L3 M4I824 Q6PQ04 C0J8Y5 P33464 A0A384V9G9 A0A384V9E4 A0A3S6JCJ2 Q9WKM6 E7E031 Q0PL00 F1CNZ1 P04135 Q5G1S6 A0A514KFM3 A0A514KFK1 S6B903 Q1W2N8 A0A1S6KDW8 D2WXM2 S6CE17 A0A1L7NS55 Q5D578 Q0ZVX1 H9TEX6 Q5G1S5 G3CI08 A0A172B6J4 I3QKR3 U3Q0B8 A0A1B4X968 Q0PL09 C6K2U0 A0A514KFL5 A0A224ANT6 A0A0K2SF30 A0A0E3GM58 Q5DQ89 A0A0K2BMP6 A0A514KFJ6 A0A1L6BQB0 P24412 D3XGJ7 A0A222UBY5 Q09PH1 A0A514KHE6 A0A514KH05 A0A514KFK0 A0A514KFM0 P09175 A0A514KFK2 Q6T524 Q64FQ9 Q7T6S9 Q19NU4 A0A088ND96 Q80RP1 C6GHD0 Q8JVR1 C5IJ46 E0AH02 C5IJ38
PDB
3Q2V
E-value=1.02704,
Score=72
Ontologies
GO
Subcellular Location
From MSLVP
Host nucleus. Host cytoplasm.
From Uniprot
Host Golgi apparatus membrane
Virion membrane
Virion membrane
Topology
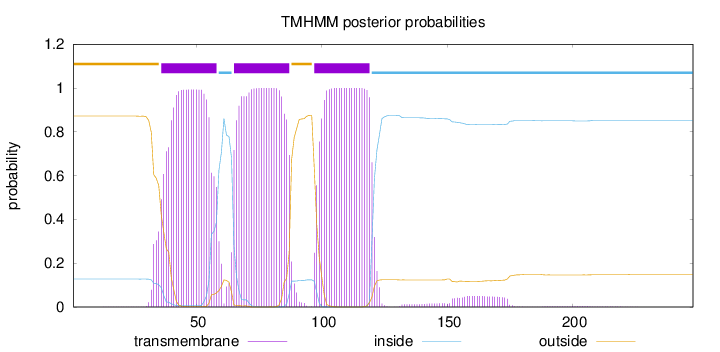
Length:
248
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
67.9104800000001
Exp number, first 60 AAs:
21.26872
Total prob of N-in:
0.12809
POSSIBLE N-term signal
sequence
outside
1 - 35
TMhelix
36 - 58
inside
59 - 64
TMhelix
65 - 87
outside
88 - 96
TMhelix
97 - 119
inside
120 - 248
Population Genetic Test Statistics
Pi
0.4411127
Theta
0.5000953
Tajima's D
-0.5129358
CLR
18.953789
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 |
Protein |
YP_009336486.1 |
|
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 |
Protein |
YP_009380524.1 |
|
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.