Strain
UK_HS_2012_NC_038294
(Region: UK; Strain: Betacoronavirus England 1, complete genome.; Date: 2012)
Gene
ORF1a protein
Description
Annotated in NCBI,
ORF1a protein
Location
GenBank Accession
Full name
Replicase polyprotein 1a
Alternative Name
ORF1a polyprotein
Sequence
CDS
ATGTCTTTCGTGGCTGGTGTGACCGCGCAAGGTGCGCGCGGTACGTATCGAGCAGCGCTCAACTCTGAAAAACATCAAGACCATGTGTCTCTAACTGTGCCACTCTGTGGTTCAGGAAACCTGGTTGAAAAACTTTCACCATGGTTCATGGATGGCGAAAATGCCTATGAAGTGGTGAAGGCCATGTTACTTAAAAAGGAGCCACTTCTCTATGTGCCCATCCGGCTGGCTGGACACACTAGACACCTCCCAGGTCCTCGTGTATACCTGGTTGAGAGGCTCATTGCTTGTGAAAATCCATTCATGGTTAACCAATTGGCTTATAGCTCTAGTGCAAATGGCAGCTTGGTTGGCACAACTTTGCAGGGCAAGCCTATTGGTATGTTCTTCCCTTATGACATCGAACTTGTCACAGGAAAGCAAAATATTCTCCTGCGCAAGTATGGCCGTGGTGGTTATCACTACACCCCATTCCACTATGAGCGAGACAACACCTCTTGCCCTGAGTGGATGGACGATTTTGAGGCGGATCCTAAAGGCAAATATGCCCAGAATCTGCTTAAGAAGTTGATTGGCGGTGATGTCACTCCAGTTGACCAATACATGTGTGGCGTTGATGGAAAACCCATTAGTGCCTACGCATTTTTAATGGCCAAGGATGGAATAACCAAACTGGCTGATGTTGAAGCGGACGTCGCAGCACGTGCTGATGACGAAGGCTTCATCACATTAAAGAACAATCTATATAGATTGGTTTGGCATGTTGAGCGTAAAGACGTTCCATATCCTAAGCAATCTATTTTTACTATTAATAGTGTGGTCCAAAAGGATGGTGTTGAAAACACTCCTCCTCACTATTTTACTCTTGGATGCAAAATTTTAACGCTCACCCCACGCAACAAGTGGAGTGGCGTTTCTGACTTGTCCCTCAAACAAAAACTCCTTTACACCTTCTATGGTAAGGAGTCACTTGAGAACCCAACCTACATTTACCACTCCGCATTCATTGAGTGTGGAAGTTGTGGTAATGATTCCTGGCTTACAGGGAATGCTATCCAAGGGTTTGCCTGTGGATGTGGGGCATCATATACAGCTAATGATGTCGAAGTCCAATCATCTGGCATGATTAAGCCAAATGCTCTTCTTTGTGCTACTTGCCCCTTTGCTAAGGGTGATAGCTGTTCTTCTAATTGCAAACATTCAGTTGCTCAGTTGGTTAGTTACCTTTCTGAACGTTGTAATGTTATTGCTGATTCTAAGTCCTTCACACTTATCTTTGGTGGCGTAGCTTACGCCTACTTTGGATGTGAGGAAGGTACTATGTACTTTGTGCCTAGAGCTAAGTCTGTTGTCTCAAGGATTGGAGACTCCATCTTTACAGGCTGTACTGGCTCTTGGAACAAGGTCACTCAAATTGCTAACATGTTCTTGGAACAGACTCAGCATTCCCTTAACTTTGTGGGAGAGTTCGTTGTCAACGATGTTGTCCTCGCAATTCTCTCTGGAACCACAACTAATGTTGACAAAATACGCCAGCTTCTCAAAGGTGTCACCCTTGACAAGTTGCGTGATTATTTAGCTGACTATGACGTAGCAGTCACTGCCGGCCCATTCATGGATAATGCTATTAATGTTGGTGGTACAGGATTACAGTATGCCGCCATTACTGCACCTTATGTAGTTCTCACTGGCTTAGGTGAGTCCTTTAAGAAAGTTGCAACCATACCGTACAAGGTTTGCAACTCTGTTAAGGATACTCTGACTTATTATGCTCACAGCGTGTTGTACAGAGTTTTTCCTTATGACATGGATTCTGGTGTGTCATCCTTTAGTGAACTACTTTTTGATTGCGTTGATCTTTCAGTAGCTTCTACCTATTTTTTAGTCCGCCTCTTGCAAGATAAGACTGGCGACTTTATGTCTACAATTATTACTTCCTGCCAAACTGCTGTTAGTAAGCTTCTAGATACATGTTTTGAAGCTACAGAAGCAACATTTAACTTCTTGTTAGATTTGGCAGGATTGTTCAGAATCTTTCTTCGCAATGCCTATGTGTACACTTCACAAGGGTTTGTGGTGGTCAATGGCAAAGTTTCTACACTTGTCAAACAAGTGTTAGACTTGCTTAATAAGGGTATGCAACTTTTGCATACAAAGGTCTCCTGGGCTGGTTCTAATATCAGTGCTGTTATCTACAGCGGCAGGGAGTCTCTAATATTCCCATCGGGAACCTATTACTGTGTCACCACTAAGGCTAAGTCCGTTCAACAAGATCTTGACGTTATTTTGCCTGGTGAGTTTTCCAAGAAGCAGTTAGGACTGCTCCAACCTACTGACAATTCTACAACTGTTAGTGTTACTGTATCCAGTAACATGGTTGAAACTGTTGTGGGTCAACTTGAGCAAACTAATATGCATAGTCCTGATGTTATAGTAGGTGACTATGTCATTATTAGTGAAAAATTGTTTGTGCGTAGTAAGGAAGAAGACGGATTTGCCTTCTACCCTGCTTGCACTAATGGTCATGCTGTACCGACTCTCTTTAGACTTAAGGGAGGTGCACCTGTAAAAAAAGTAGCCTTTGGCGGTGATCAAGTACATGAGGTTGCTGCTGTAAGAAGTGTTACTGTCGAGTACAACATTCATGCTGTATTAGACACACTACTTGCTTCTTCTAGTCTTAGAACCTTTGTTGTAGATAAGTCTTTGTCAATTGAGGAGTTTGCTGACGTAGTAAAGGAACAAGTCTCAGACTTGCTTGTTAAATTACTGCGTGGAATGCCGATTCCAGATTTTGATTTAGACGATTTTATTGACGCACCATGCTATTGCTTTAACGCTGAGGGTGATGCATCTTGGTCTTCTACTATGATCTTCTCTCTTCACCCCGTCGAGTGTGACGAGGAGTGTTCTGAAGTAGAGGCTTCAGATTTAGAAGAAGGTGAATCAGAGTGCATTTCTGAGACTTCAACTGAACAAGTTGACGTTTCTCATGAGATTTCTGACGACGAGTGGGCTGCTGCAGTTGATGAAGCGTTCCCCCTCGATGAAGCAGAAGATGTTACTGAATCTGTGCAAGAAGAAGCACAACCAGTAGAAGTACCTGTTGAAGATATTGCGCAGGTTGTCATAGCTGACACCTTACAGGAAACTCCTGTTGTGTCTGATACTGTTGAAGTCCCACCGCAAGTGGTGAAACTTCCGTCTGAACCTCAGACTATCCAGCCCGAGGTAAAAGAAGTTGCACCTGTCTATGAGGCTGATACCGAACAGACACAGAGTGTTACTGTTAAACCTAAGAGGTTACGCAAAAAGCGTAATGTTGACCCTTTGTCCAATTTTGAACATAAGGTTATTACAGAGTGCGTTACCATAGTTTTAGGTGACGCAATTCAAGTAGCCAAGTGCTATGGGGAGTCTGTGTTAGTTAATGCTGCTAACACACATCTTAAGCATGGCGGTGGTATCGCTGGTGCTATTAATGCGGCTTCAAAAGGGGCTGTCCAAAAAGAGTCAGATGAGTATATTCTGGCTAAAGGGCCGTTACAAGTAGGAGATTCAGTTCTCTTGCAAGGCCATTCTCTAGCTAAGAATATCCTGCATGTCGTAGGCCCAGATGCCCGCGCTAAACAGGATGTTTCTCTCCTTAGTAAGTGCTATAAGGCTATGAATGCATATCCTCTTGTAGTCACTCCTCTTGTTTCAGCAGGCATATTTGGTGTAAAACCAGCTGTGTCTTTTGATTATCTTATTAGAGAGGCTAAGACTAGAGTTTTAGTCGTCGTTAATTCCCAAGATGTCTATAAGAGTCTTACCATAGTTGACATTCCACAGAGTTTGACTTTTTCATATGATGGGTTACGTGGCGCAATACGTAAAGCTAAAGATTATGGTTTTACTGTTTTTGTGTGCACAGACAACTCTGCTAACACTAAAGTTCTTAGGAACAAGGGTGTTGATTATACTAAGAAGTTTCTTACAGTTGACGGTGTGCAATATTATTGCTACACGTCTAAGGACACTTTAGATGATATCTTACAACAGGCTAATAAGTCTGTTGGTATTATATCTATGCCTTTGGGATATGTGTCTCATGGTTTAGACTTAATTCAAGCAGGGAGTGTCGTGCGTAGAGTTAACGTGCCCTACGTGTGTCTCCTAGCTAATAAAGAGCAAGAAGCTATTTTGATGTCTGAAGACGTTAAGTTAAACCCTTCAGAAGATTTTATAAAGCACGTCCGCACTAATGGTGGTTACAATTCTTGGCATTTAGTCGAGGGTGAACTATTGGTGCAAGACTTACGCTTAAATAAGCTCCTGCATTGGTCTGATCAAACCATATGCTACAAGGATAGTGTGTTTTATGTTGTAAAGAATAGTACAGCTTTTCCATTTGAAACACTTTCAGCATGTCGTGCGTATTTGGATTCACGCACGACACAGCAGTTAACAATCGAAGTCTTAGTGACTGTCGATGGTGTAAATTTTAGAACAGTCGTTCTAAATAATAAGAACACTTATAGATCACAGCTTGGATGCGTTTTCTTTAATGGTGCTGATATTTCTGATACCATTCCTGATGAGAAACAGAATGGTCACAGTTTATATCTAGCAGACAATTTGACTGCTGATGAAACAAAGGCGCTTAAAGAGTTATATGGCCCCGTTGATCCTACTTTCTTACACAGATTCTATTCACTTAAGGCTGCAGTCCATAAGTGGAAGATGGTTGTGTGTGATAAGGTACGTTCTCTCAAATTGAGTGATAATAATTGTTATCTTAATGCAGTTATTATGACACTTGATTTATTGAAGGACATTAAATTTGTTATACCTGCTCTACAGCATGCATTTATGAAACATAAGGGCGGTGATTCAACTGACTTCATAGCCCTCATTATGGCTTATGGCAATTGCACATTTGGTGCTCCAGATGATGCCTCTCGGTTACTTCATACCGTGCTTGCAAAGGCTGAGTTATGCTGTTCTGCACGCATGGTTTGGAGAGAGTGGTGCAATGTCTGTGGCATAAAAGATGTTGTTCTACAAGGCTTAAAAGCTTGTTGTTACGTGGGTGTGCAAACTGTTGAAGATCTGCGTGCTCGCATGACATATGTATGCCAGTGTGGTGGTGAACGTCATCGGCAAATAGTCGAACACACCACCCCCTGGTTGCTGCTCTCAGGCACACCAAATGAAAAATTGGTGACAACCTCCACGGCGCCTGATTTTGTAGCGTTTAATGTCTTTCAGGGCATTGAAACGGCTGTTGGCCATTATGTTCATGCTCGCCTGAAGGGTGGTCTTATTTTAAAGTTTGACTCTGGCACCGTTAGCAAGACTTCAGACTGGAAGTGCAAGGTGACAGATGTACTTTTCCCCGGCCAAAAATACAGTAGCGATTGTAATGTCGTACGGTATTCTTTGGACGGTAATTTCAGAACAGAGGTTGATCCCGACCTATCTGCTTTCTATGTTAAGGATGGTAAATACTTTACAAGTGAACCACCCGTAACATATTCACCAGCTACAATTTTAGCTGGTAGTGTCTACACTAATAGCTGCCTTGTATCGTCTGATGGACAACCTGGCGGTGATGCTATTAGTTTGAGTTTTAATAACCTTTTAGGGTTTGATTCTAGTAAACCAGTCACTAAGAAATACACTTACTCCTTCTTGCCTAAAGAAGACGGCGATGTGTTGTTGGCTGAGTTTGACACTTATGACCCTATTTATAAGAATGGTGCCATGTATAAAGGCAAACCAATTCTTTGGGTCAACAAAGCATCTTATGATACTAATCTTAATAAGTTCAATAGAGCTAGTTTGCGTCAAATTTTTGACGTAGCCCCCATTGAACTCGAAAATAAATTCACACCTTTGAGTGTGGAGTCTACACCAGTTGAACCTCCAACTGTAGATGTGGTAGCACTTCAACAGGAAATGACAATTGTCAAATGTAAGGGTTTAAATAAACCTTTCGTGAAGGACAATGTCAGTTTCGTTGCTGATGACTCAGGTACTCCCGTTGTTGAGTATCTGTCTAAAGAAGATCTACATACATTGTATGTAGACCCTAAGTATCAAGTCATTGTCTTAAAAGACAATGTACTTTCTTCTATGCTTAGATTGCACACCGTTGAGTCAGGTGATATTAACGTTGTTGCAGCTTCCGGATCTTTGACACGTAAAGTGAAGTTACTATTTAGGGCTTCATTTTATTTCAAAGAATTTGCTACCCGCACTTTCACTGCTACCACTGCTGTAGGTAGTTGTATAAAGAGTGTAGTGCGGCATCTAGGTGTTACTAAAGGCATATTGACAGGCTGTTTTAGTTTTGTCAAGATGTTATTTATGCTTCCACTAGCTTACTTTAGTGATTCAAAACTCGGCACCACAGAGGTTAAAGTGAGTGCTTTGAAAACAGCTGGCGTTGTGACAGGTAATGTTGTAAAACAGTGTTGCACTGCTGCTGTTGATTTAAGTATGGATAAGTTGCGCCGTGTGGATTGGAAATCAACCCTACGGTTGTTACTTATGTTATGCACAACTATGGTATTGTTGTCTTCTGTGTATCACTTGTATGTCTTCAATCAGGTCTTATCAAGTGATGTTATGTTTGAAGATGCCCAAGGTTTGAAAAAGTTCTACAAAGAAGTTAGAGCTTACCTAGGAATCTCTTCTGCTTGTGACGGTCTTGCTTCAGCTTATAGGGCGAATTCCTTTGATGTACCTACATTCTGCGCAAACCGTTCTGCAATGTGTAATTGGTGCTTGATTAGCCAAGATTCCATAACTCACTACCCAGCTCTTAAGATGGTTCAAACACATCTTAGCCACTATGTTCTTAACATAGATTGGTTGTGGTTTGCATTTGAGACTGGTTTGGCATACATGCTCTATACCTCGGCCTTCAACTGGTTGTTGTTGGCAGGTACATTGCATTATTTCTTTGCACAGACTTCCATATTTGTAGACTGGCGGTCATACAATTATGCTGTGTCTAGTGCCTTCTGGTTATTCACCCACATTCCAATGGCGGGTTTGGTACGAATGTATAATTTGTTAGCATGCCTTTGGCTTTTACGCAAGTTTTATCAGCATGTAATCAATGGTTGCAAAGATACGGCATGCTTGCTCTGCTATAAGAGGAACCGACTTACTAGAGTTGAAGCTTCTACCGTTGTCTGTGGTGGAAAACGTACGTTTTATATCACAGCAAATGGCGGTATTTCATTCTGTCGTAGGCATAATTGGAATTGTGTGGATTGTGACACTGCAGGTGTGGGGAATACCTTCATCTGTGAAGAAGTCGCAAATGACCTCACTACCGCCCTACGCAGGCCTATTAACGCTACGGATAGATCACATTATTATGTGGATTCCGTTACAGTTAAAGAGACTGTTGTTCAGTTTAATTATCGTAGAGACGGTCAACCATTCTACGAGCGGTTTCCCCTCTGCGCTTTTACAAATCTAGATAAGTTGAAGTTCAAAGAGGTCTGTAAAACTACTACTGGTATACCTGAATACAACTTTATCATCTACGACTCATCAGATCGTGGCCAGGAAAGTTTAGCTAGGTCTGCATGTGTTTATTATTCTCAAGTCTTGTGTAAATCAATTCTTTTGGTTGACTCAAGTTTGGTTACTTCTGTTGGTGATTCTAGTGAAATCGCCACTAAAATGTTTGATTCCTTTGTTAATAGTTTCGTCTCGCTGTATAATGTCACACGCGATAAGTTGGAAAAACTTATCTCTACTGCTCGTGATGGCGTAAGGCGAGGCGATAACTTCCATAGTGTCTTAACAACATTCATTGACGCAGCACGAGGCCCCGCAGGTGTGGAGTCTGATGTTGAGACCAATGAAATTGTTGACTCTGTGCAGTATGCTCATAAACATGACATACAAATTACTAATGAGAGTTACAATAATTATGTACCCTCATATGTTAAACCTGATAGTGTGTCTACCAGTGATTTAGGTAGTCTCATTGATTGTAATGCGGCTTCAGTTAACCAAATTGTCTTGCGTAATTCTAATGGTGCTTGTATTTGGAACGCTGCTGCATATATGAAACTCTCGGATGCACTTAAACGACAGATTCGCATTGCATGCCGTAAGTGTAATTTAGCTTTCCGGTTAACCACCTCAAAGCTACGCGCTAATGATAATATCTTATCAGTTAGATTCACTGCTAACAAAATTGTTGGTGGTGCTCCTACATGGTTTAATGCGTTGCGTGACTTTACGTTAAAGGGTTACGTTCTTGCTACCATTATTGTGTTTCTGTGTGCTGTACTGATGTATTTGTGTTTACCTACATTTTCTATGGTACCTGTTGAATTTTATGAAGACCGCATCTTGGACTTTAAAGTTCTTGATAATGGTATCATTAGGGATGTAAATCCTGATGATAAGTGCTTTGCTAATAAGCACCGGTCCTTCACACAATGGTATCATGAGCATGTTGGTGGTGTCTATGACAACTCTATCACATGCCCATTGACAGTTGCAGTAATTGCTGGAGTTGCTGGTGCTCGCATTCCAGACGTACCTACTACATTGGCTTGGGTGAACAATCAGATAATTTTCTTTGTTTCTCGAGTCTTTGCTAATACAGGCAGTGTTTGCTACACTCCTATAGATGAGATACCCTATAAGAGTTTCTCTGATAGTGGTTGCATTCTTCCATCTGAGTGCACTATGTTTAGGGATGCAGAGGGCCGTATGACACCATACTGCCATGATCCTACTGTTTTGCCTGGGGCTTTTGCGTACAGTCAGATGAGGCCTCATGTTCGTTACGACTTGTATGATGGTAACATGTTTATTAAATTTCCTGAAGTAGTATTTGAAAGTACACTTAGGATTACTAGAACTCTGTCAACTCAGTACTGCCGGTTCGGTAGTTGTGAGTATGCACAAGAGGGTGTTTGTATTACCACAAATGGCTCGTGGGCCATTTTTAATGACCACCATCTTAATAGACCTGGTGTCTATTGTGGCTCTGATTTTATTGACATTGTCAGGCGGTTAGCAGTATCACTGTTCCAGCCTATTACTTATTTCCAATTGACTACCTCATTGGTCTTGGGTATAGGTTTGTGTGCGTTCCTGACTTTGCTCTTCTATTATATTAATAAAGTAAAACGTGCTTTTGCAGATTACACCCAGTGTGCTGTAATTGCTGTTGTTGCTGCTGTTCTTAATAGCTTGTGCATCTGCTTTGTTGCCTCTATACCATTGTGTATAGTACCTTACACTGCATTGTACTATTATGCTACATTCTATTTTACTAATGAGCCTGCATTTATTATGCATGTTTCTTGGTACATTATGTTCGGGCCTATCGTTCCCATATGGATGACCTGCGTCTATACAGTTGCAATGTGCTTTAGACACTTCTTCTGGGTTTTAGCTTATTTTAGTAAGAAACATGTAGAAGTTTTTACTGATGGTAAGCTTAATTGTAGTTTCCAGGACGCTGCCTCTAATATCTTTGTTATTAACAAGGACACTTATGCAGCTCTTAGAAACTCTTTAACTAATGATGCCTATTCACGATTTTTGGGGTTGTTTAACAAGTATAAGTACTTCTCTGGTGCTATGGAAACAGCCGCTTATCGTGAAGCTGCAGCATGTCATCTTGCTAAAGCCTTACAAACATACAGCGAGACTGGTAGTGATCTTCTTTACCAACCACCCAACTGTAGCATAACCTCTGGCGTGTTGCAAAGCGGTTTGGTGAAAATGTCACATCCCAGTGGAGATGTTGAGGCTTGTATGGTTCAGGTTACCTGCGGTAGCATGACTCTTAATGGTCTTTGGCTTGACAACACAGTCTGGTGCCCACGACACGTAATGTGCCCGGCTGACCAGTTGTCTGATCCTAATTATGATGCCTTGTTGATTTCTATGACTAATCATAGTTTCAGTGTGCAAAAACACATTGGCGCTCCAGCAAACTTGCGTGTTGTTGGTCATGCCATGCAAGGCACTCTTTTGAAGTTGACTGTCGATGTTGCTAACCCTAGCACTCCAGCCTACACTTTTACAACAGTGAAACCTGGCGCAGCATTTAGTGTGTTAGCATGCTATAATGGTCGTCCGACTGGTACATTCACTGTTGTAATGCGCCCTAACTACACAATTAAGGGTTCCTTTCTGTGTGGTTCTTGTGGTAGTGTTGGTTACACCAAGGAGGGTAGTGTGATCAATTTTTGTTACATGCATCAAATGGAACTTGCTAATGGTACACATACCGGTTCAGCATTTGATGGTACTATGTATGGTGCCTTTATGGATAAACAAGTGCACCAAGTTCAGTTAACAGACAAATACTGCAGTGTTAATGTAGTAGCTTGGCTTTACGCAGCAATACTTAATGGTTGCGCTTGGTTTGTAAAACCTAATCGCACTAGTGTTGTTTCTTTTAATGAATGGGCTCTTGCCAACCAATTCACTGAATTTGTTGGCACTCAATCCGTTGACATGTTAGCTGTCAAAACAGGCGTTGCTATTGAACAGCTGCTTTATGCGATCCAACAACTTTATACTGGGTTCCAGGGAAAGCAAATCCTTGGCAGTACCATGTTGGAAGATGAATTCACACCTGAGGATGTTAATATGCAGATTATGGGTGTGGTTATGCAGAGTGGTGTGAGAAAAGTTACATATGGTACTGCGCATTGGTTGTTCGCGACCCTTGTCTCAACCTATGTGATAATCTTACAAGCCACTAAATTTACTTTGTGGAACTACTTGTTTGAGACTATTCCCACACAGTTGTTCCCACTCTTATTTGTGACTATGGCCTTCGTTATGTTGTTGGTTAAACACAAACACACCTTTTTGACACTTTTCTTGTTGCCTGTGGCTATTTGTTTGACTTATGCAAACATAGTCTACGAGCCCACTACTCCCATTTCGTCAGCGCTGATTGCAGTTGCAAATTGGCTTGCCCCCACTAATGCTTATATGCGCACTACACATACTGATATTGGTGTCTACATTAGTATGTCACTTGTATTAGTCATTGTAGTGAAGAGATTGTACAACCCATCACTTTCTAACTTTGCGTTAGCATTGTGCAGTGGTGTAATGTGGTTGTACACTTATAGCATTGGAGAAGCCTCAAGCCCCATTGCCTATCTGGTTTTTGTCACTACACTCACTAGTGATTATACGATTACAGTCTTTGTTACTGTTAACCTTGCAAAAGTTTGCACTTATGCCATCTTTGCTTACTCGCCACAGCTTACACTTGTGTTTCCGGAAGTGAAGATGATACTTTTATTATACACATGTTTAGGTTTCATGTGTACTTGCTATTTTGGTGTCTTCTCTCTTTTGAACCTTAAGCTTAGAGCACCTATGGGTGTCTATGACTTTAAGGTCTCAACACAAGAGTTCAGATTCATGACAGCTAACAATCTAACTGCACCTAGAAATTCTTGGGAGGCTATGGCTCTGAACTTTAAGTTAATAGGTATTGGCGGTACACCTTGTATAAAGGTTGCTGCTATGCAGTCTAAACTTACAGATCTTAAATGCACATCTGTGGTTCTCCTCTCTGTGCTCCAACAGTTACACTTAGAGGCTAATAGTAGGGCCTGGGCTTTCTGTGTTAAATGCCATAATGATATATTGGCAGCAACAGACCCCAGTGAGGCTTTCGAGAAATTCGTAAGTCTCTTTGCCACTTTAATGACTTTTTCTGGTAATGTAGATCTTGATGCGTTAGCTAGTGATATTTTTGACACTCCTAGCGTACTTCAAGCTACTCTTTCTGAGTTTTCACACTTAGCTACCTTTGCTGAGTTGGAAGCTGCGCAGAAAGCCTATCAGGAAGCTATGGACTCTGGTGACACCTCACCACAAGTTCTTAAGGCTTTGCAGAAGGCTGTTAATATAGCTAAAAACGCCTATGAGAAGGATAAGGCAGTGGCCCGTAAGTTAGAACGTATGGCTGATCAGGCTATGACTTCTATGTATAAGCAAGCACGTGCTGAAGACAAGAAAGCAAAAATTGTCAGTGCTATGCAAACTATGTTGTTTGGTATGATTAAGAAGCTCGACAACGATGTTCTTAATGGTATCATTTCTAACGCTAGGAATGGTTGTATACCTCTTAGTGTCATTCCACTGTGTGCTTCAAATAAACTTCGCGTTGTAATTCCTGACTTCACCGTCTGGAATCAGGTAGTCACATATCCCTCGCTTAACTACGCTGGGGCTTTGTGGGACATTACAGTTATAAACAATGTGGACAATGAAATTGTTAAGTCTTCAGATGTTGTAGACAGCAATGAAAATTTAACATGGCCACTTGTTTTAGAATGCACTAGGGCATCCACTTCTGCCGTTAAGTTGCAAAATAATGAGATCAAACCTTCAGGTTTAAAAACCATGGTTGTGTCTGCAGGTCAAGAGCAAACTAACTGTAATACTAGTTCCTTAGCTTATTACGAACCTGTGCAGGGTCGTAAAATGCTGATGGCTCTTCTTTCTGATAATGCCTATCTCAAATGGGCGCGTGTTGAAGGTAAGGACGGATTTGTTAGTGTAGAGCTACAACCTCCTTGCAAATTCTTGATTGCGGGACCAAAAGGACCTGAAATCCGATATCTCTATTTTGTTAAAAATCTTAACAACCTTCATCGCGGGCAAGTGTTAGGGCACATTGCTGCGACTGTTAGATTGCAAGCTGGTTCTAACACCGAGTTTGCCTCTAATTCTTCGGTGTTGTCACTTGTTAACTTCACCGTTGATCCTCAAAAAGCTTATCTCGATTTCGTCAATGCGGGAGGTGCCCCATTGACAAATTGTGTTAAGATGCTTACTCCTAAAACTGGTACAGGTATAGCTATATCTGTTAAACCAGAGAGTACAGCTGATCAAGAGACTTATGGTGGAGCTTCAGTGTGTCTCTATTGCCGTGCGCATATAGAACATCCTGATGTCTCTGGTGTTTGTAAATATAAGGGTAAGTTTGTCCAAATCCCTGCTCAGTGTGTCCGTGACCCTGTGGGATTTTGTTTGTCAAATACCCCCTGTAATGTCTGTCAATATTGGATTGGATATGGGTGCAATTGTGACTCGCTTAGGCAAGCAGCACTGCCCCAATCTAAAGATTCCAATTTTTTAAACGAGTCCGGGGTTCTATTGTAA
Protein
MSFVAGVTAQGARGTYRAALNSEKHQDHVSLTVPLCGSGNLVEKLSPWFMDGENAYEVVKAMLLKKEPLLYVPIRLAGHTRHLPGPRVYLVERLIACENPFMVNQLAYSSSANGSLVGTTLQGKPIGMFFPYDIELVTGKQNILLRKYGRGGYHYTPFHYERDNTSCPEWMDDFEADPKGKYAQNLLKKLIGGDVTPVDQYMCGVDGKPISAYAFLMAKDGITKLADVEADVAARADDEGFITLKNNLYRLVWHVERKDVPYPKQSIFTINSVVQKDGVENTPPHYFTLGCKILTLTPRNKWSGVSDLSLKQKLLYTFYGKESLENPTYIYHSAFIECGSCGNDSWLTGNAIQGFACGCGASYTANDVEVQSSGMIKPNALLCATCPFAKGDSCSSNCKHSVAQLVSYLSERCNVIADSKSFTLIFGGVAYAYFGCEEGTMYFVPRAKSVVSRIGDSIFTGCTGSWNKVTQIANMFLEQTQHSLNFVGEFVVNDVVLAILSGTTTNVDKIRQLLKGVTLDKLRDYLADYDVAVTAGPFMDNAINVGGTGLQYAAITAPYVVLTGLGESFKKVATIPYKVCNSVKDTLTYYAHSVLYRVFPYDMDSGVSSFSELLFDCVDLSVASTYFLVRLLQDKTGDFMSTIITSCQTAVSKLLDTCFEATEATFNFLLDLAGLFRIFLRNAYVYTSQGFVVVNGKVSTLVKQVLDLLNKGMQLLHTKVSWAGSNISAVIYSGRESLIFPSGTYYCVTTKAKSVQQDLDVILPGEFSKKQLGLLQPTDNSTTVSVTVSSNMVETVVGQLEQTNMHSPDVIVGDYVIISEKLFVRSKEEDGFAFYPACTNGHAVPTLFRLKGGAPVKKVAFGGDQVHEVAAVRSVTVEYNIHAVLDTLLASSSLRTFVVDKSLSIEEFADVVKEQVSDLLVKLLRGMPIPDFDLDDFIDAPCYCFNAEGDASWSSTMIFSLHPVECDEECSEVEASDLEEGESECISETSTEQVDVSHEISDDEWAAAVDEAFPLDEAEDVTESVQEEAQPVEVPVEDIAQVVIADTLQETPVVSDTVEVPPQVVKLPSEPQTIQPEVKEVAPVYEADTEQTQSVTVKPKRLRKKRNVDPLSNFEHKVITECVTIVLGDAIQVAKCYGESVLVNAANTHLKHGGGIAGAINAASKGAVQKESDEYILAKGPLQVGDSVLLQGHSLAKNILHVVGPDARAKQDVSLLSKCYKAMNAYPLVVTPLVSAGIFGVKPAVSFDYLIREAKTRVLVVVNSQDVYKSLTIVDIPQSLTFSYDGLRGAIRKAKDYGFTVFVCTDNSANTKVLRNKGVDYTKKFLTVDGVQYYCYTSKDTLDDILQQANKSVGIISMPLGYVSHGLDLIQAGSVVRRVNVPYVCLLANKEQEAILMSEDVKLNPSEDFIKHVRTNGGYNSWHLVEGELLVQDLRLNKLLHWSDQTICYKDSVFYVVKNSTAFPFETLSACRAYLDSRTTQQLTIEVLVTVDGVNFRTVVLNNKNTYRSQLGCVFFNGADISDTIPDEKQNGHSLYLADNLTADETKALKELYGPVDPTFLHRFYSLKAAVHKWKMVVCDKVRSLKLSDNNCYLNAVIMTLDLLKDIKFVIPALQHAFMKHKGGDSTDFIALIMAYGNCTFGAPDDASRLLHTVLAKAELCCSARMVWREWCNVCGIKDVVLQGLKACCYVGVQTVEDLRARMTYVCQCGGERHRQIVEHTTPWLLLSGTPNEKLVTTSTAPDFVAFNVFQGIETAVGHYVHARLKGGLILKFDSGTVSKTSDWKCKVTDVLFPGQKYSSDCNVVRYSLDGNFRTEVDPDLSAFYVKDGKYFTSEPPVTYSPATILAGSVYTNSCLVSSDGQPGGDAISLSFNNLLGFDSSKPVTKKYTYSFLPKEDGDVLLAEFDTYDPIYKNGAMYKGKPILWVNKASYDTNLNKFNRASLRQIFDVAPIELENKFTPLSVESTPVEPPTVDVVALQQEMTIVKCKGLNKPFVKDNVSFVADDSGTPVVEYLSKEDLHTLYVDPKYQVIVLKDNVLSSMLRLHTVESGDINVVAASGSLTRKVKLLFRASFYFKEFATRTFTATTAVGSCIKSVVRHLGVTKGILTGCFSFVKMLFMLPLAYFSDSKLGTTEVKVSALKTAGVVTGNVVKQCCTAAVDLSMDKLRRVDWKSTLRLLLMLCTTMVLLSSVYHLYVFNQVLSSDVMFEDAQGLKKFYKEVRAYLGISSACDGLASAYRANSFDVPTFCANRSAMCNWCLISQDSITHYPALKMVQTHLSHYVLNIDWLWFAFETGLAYMLYTSAFNWLLLAGTLHYFFAQTSIFVDWRSYNYAVSSAFWLFTHIPMAGLVRMYNLLACLWLLRKFYQHVINGCKDTACLLCYKRNRLTRVEASTVVCGGKRTFYITANGGISFCRRHNWNCVDCDTAGVGNTFICEEVANDLTTALRRPINATDRSHYYVDSVTVKETVVQFNYRRDGQPFYERFPLCAFTNLDKLKFKEVCKTTTGIPEYNFIIYDSSDRGQESLARSACVYYSQVLCKSILLVDSSLVTSVGDSSEIATKMFDSFVNSFVSLYNVTRDKLEKLISTARDGVRRGDNFHSVLTTFIDAARGPAGVESDVETNEIVDSVQYAHKHDIQITNESYNNYVPSYVKPDSVSTSDLGSLIDCNAASVNQIVLRNSNGACIWNAAAYMKLSDALKRQIRIACRKCNLAFRLTTSKLRANDNILSVRFTANKIVGGAPTWFNALRDFTLKGYVLATIIVFLCAVLMYLCLPTFSMVPVEFYEDRILDFKVLDNGIIRDVNPDDKCFANKHRSFTQWYHEHVGGVYDNSITCPLTVAVIAGVAGARIPDVPTTLAWVNNQIIFFVSRVFANTGSVCYTPIDEIPYKSFSDSGCILPSECTMFRDAEGRMTPYCHDPTVLPGAFAYSQMRPHVRYDLYDGNMFIKFPEVVFESTLRITRTLSTQYCRFGSCEYAQEGVCITTNGSWAIFNDHHLNRPGVYCGSDFIDIVRRLAVSLFQPITYFQLTTSLVLGIGLCAFLTLLFYYINKVKRAFADYTQCAVIAVVAAVLNSLCICFVASIPLCIVPYTALYYYATFYFTNEPAFIMHVSWYIMFGPIVPIWMTCVYTVAMCFRHFFWVLAYFSKKHVEVFTDGKLNCSFQDAASNIFVINKDTYAALRNSLTNDAYSRFLGLFNKYKYFSGAMETAAYREAAACHLAKALQTYSETGSDLLYQPPNCSITSGVLQSGLVKMSHPSGDVEACMVQVTCGSMTLNGLWLDNTVWCPRHVMCPADQLSDPNYDALLISMTNHSFSVQKHIGAPANLRVVGHAMQGTLLKLTVDVANPSTPAYTFTTVKPGAAFSVLACYNGRPTGTFTVVMRPNYTIKGSFLCGSCGSVGYTKEGSVINFCYMHQMELANGTHTGSAFDGTMYGAFMDKQVHQVQLTDKYCSVNVVAWLYAAILNGCAWFVKPNRTSVVSFNEWALANQFTEFVGTQSVDMLAVKTGVAIEQLLYAIQQLYTGFQGKQILGSTMLEDEFTPEDVNMQIMGVVMQSGVRKVTYGTAHWLFATLVSTYVIILQATKFTLWNYLFETIPTQLFPLLFVTMAFVMLLVKHKHTFLTLFLLPVAICLTYANIVYEPTTPISSALIAVANWLAPTNAYMRTTHTDIGVYISMSLVLVIVVKRLYNPSLSNFALALCSGVMWLYTYSIGEASSPIAYLVFVTTLTSDYTITVFVTVNLAKVCTYAIFAYSPQLTLVFPEVKMILLLYTCLGFMCTCYFGVFSLLNLKLRAPMGVYDFKVSTQEFRFMTANNLTAPRNSWEAMALNFKLIGIGGTPCIKVAAMQSKLTDLKCTSVVLLSVLQQLHLEANSRAWAFCVKCHNDILAATDPSEAFEKFVSLFATLMTFSGNVDLDALASDIFDTPSVLQATLSEFSHLATFAELEAAQKAYQEAMDSGDTSPQVLKALQKAVNIAKNAYEKDKAVARKLERMADQAMTSMYKQARAEDKKAKIVSAMQTMLFGMIKKLDNDVLNGIISNARNGCIPLSVIPLCASNKLRVVIPDFTVWNQVVTYPSLNYAGALWDITVINNVDNEIVKSSDVVDSNENLTWPLVLECTRASTSAVKLQNNEIKPSGLKTMVVSAGQEQTNCNTSSLAYYEPVQGRKMLMALLSDNAYLKWARVEGKDGFVSVELQPPCKFLIAGPKGPEIRYLYFVKNLNNLHRGQVLGHIAATVRLQAGSNTEFASNSSVLSLVNFTVDPQKAYLDFVNAGGAPLTNCVKMLTPKTGTGIAISVKPESTADQETYGGASVCLYCRAHIEHPDVSGVCKYKGKFVQIPAQCVRDPVGFCLSNTPCNVCQYWIGYGCNCDSLRQAALPQSKDSNFLNESGVLL
Summary
Function
The replicase polyprotein of coronaviruses is a multifunctional protein: it contains the activities necessary for the transcription of negative stranded RNA, leader RNA, subgenomic mRNAs and progeny virion RNA as well as proteinases responsible for the cleavage of the polyprotein into functional products.
Promotes the degradation of host mRNAs by inducing an endonucleolytic RNA cleavage in template mRNAs, and inhibits of host mRNA translation, a function that is separable from its RNA cleavage activity. By suppressing host gene expression, nsp1 facilitates efficient viral gene expression in infected cells and evasion from host immune response.
May play a role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2. Indeed, these two proteins play a role in maintaining the functional integrity of the mitochondria and protecting cells from various stresses.
Responsible for the cleavages located at the N-terminus of the replicase polyprotein. In addition, PL-PRO possesses a deubiquitinating/deISGylating activity and processes both 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains from cellular substrates. Participates together with nsp4 in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication. Antagonizes innate immune induction of type I interferon by blocking the phosphorylation, dimerization and subsequent nuclear translocation of host IRF3. Prevents also host NF-kappa-B signaling.
Participates in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication.
Proteinase 3CL-PRO: Cleaves the C-terminus of replicase polyprotein at 11 sites. Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. Also able to bind an ADP-ribose-1''-phosphate (ADRP).
Plays a role in the initial induction of autophagosomes from host reticulum endoplasmic. Later, limits the expansion of these phagosomes that are no longer able to deliver viral components to lysosomes.
Forms a hexadecamer with nsp8 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
Forms a hexadecamer with nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
May participate in viral replication by acting as a ssRNA-binding protein.
Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease and nsp16 2'-O-methyltransferase activities. Therefore plays an essential role in viral mRNAs cap methylation.
Promotes the degradation of host mRNAs by inducing an endonucleolytic RNA cleavage in template mRNAs, and inhibits of host mRNA translation, a function that is separable from its RNA cleavage activity. By suppressing host gene expression, nsp1 facilitates efficient viral gene expression in infected cells and evasion from host immune response.
May play a role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2. Indeed, these two proteins play a role in maintaining the functional integrity of the mitochondria and protecting cells from various stresses.
Responsible for the cleavages located at the N-terminus of the replicase polyprotein. In addition, PL-PRO possesses a deubiquitinating/deISGylating activity and processes both 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains from cellular substrates. Participates together with nsp4 in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication. Antagonizes innate immune induction of type I interferon by blocking the phosphorylation, dimerization and subsequent nuclear translocation of host IRF3. Prevents also host NF-kappa-B signaling.
Participates in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication.
Proteinase 3CL-PRO: Cleaves the C-terminus of replicase polyprotein at 11 sites. Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. Also able to bind an ADP-ribose-1''-phosphate (ADRP).
Plays a role in the initial induction of autophagosomes from host reticulum endoplasmic. Later, limits the expansion of these phagosomes that are no longer able to deliver viral components to lysosomes.
Forms a hexadecamer with nsp8 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
Forms a hexadecamer with nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
May participate in viral replication by acting as a ssRNA-binding protein.
Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease and nsp16 2'-O-methyltransferase activities. Therefore plays an essential role in viral mRNAs cap methylation.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
TSAVLQ-|-SGFRK-NH(2) and SGVTFQ-|-GKFKK the two peptides corresponding to the two self-cleavage sites of the SARS 3C-like proteinase are the two most reactive peptide substrates. The enzyme exhibits a strong preference for substrates containing Gln at P1 position and Leu at P2 position.
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
TSAVLQ-|-SGFRK-NH(2) and SGVTFQ-|-GKFKK the two peptides corresponding to the two self-cleavage sites of the SARS 3C-like proteinase are the two most reactive peptide substrates. The enzyme exhibits a strong preference for substrates containing Gln at P1 position and Leu at P2 position.
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Subunit
Nsp2 interacts with host PHB and PHB2. 3CL-PRO exists as monomer and homodimer. Nsp4 interacts with PL-PRO and nsp6. Only the homodimer shows catalytic activity. Eight copies of nsp7 and eight copies of nsp8 assemble to form a heterohexadecamer dsRNA-encircling ring structure. Nsp9 is a dimer.
Miscellaneous
Produced by conventional translation.
Similarity
Belongs to the coronaviruses polyprotein 1ab family.
Keywords
3D-structure
Activation of host autophagy by virus
Decay of host mRNAs by virus
Eukaryotic host gene expression shutoff by virus
Eukaryotic host translation shutoff by virus
Host cytoplasm
Host gene expression shutoff by virus
Host membrane
Host mRNA suppression by virus
Host-virus interaction
Hydrolase
Inhibition of host innate immune response by virus
Inhibition of host interferon signaling pathway by virus
Inhibition of host IRF3 by virus
Inhibition of host ISG15 by virus
Inhibition of host RLR pathway by virus
Membrane
Metal-binding
Modulation of host ubiquitin pathway by viral deubiquitinase
Modulation of host ubiquitin pathway by virus
Protease
Reference proteome
Repeat
Ribosomal frameshifting
RNA-binding
Thiol protease
Transmembrane
Transmembrane helix
Ubl conjugation pathway
Viral immunoevasion
Zinc
Zinc-finger
Feature
chain Host translation inhibitor nsp1
Uniprot
EMBL
Proteomes
PRIDE
Pfam
Interpro
IPR024375
Nsp3_coronavir
IPR037230 NSP8_sf
IPR018995 RNA_synth_NSP10_coronavirus
IPR037204 NSP7_sf
IPR014827 Viral_protease
IPR038123 NSP4_C_sf
IPR008740 Peptidase_C30
IPR014828 NSP7
IPR032505 Corona_NSP4_C
IPR014822 NSP9
IPR032592 NAR_dom
IPR002589 Macro_dom
IPR009003 Peptidase_S1_PA
IPR036499 NSP9_sf
IPR013016 Peptidase_C30/C16
IPR038400 Nsp3_coronavir_sf
IPR014829 NSP8
IPR036333 NSP10_sf
IPR042570 NAR_sf
IPR038083 R1a/1ab
IPR037230 NSP8_sf
IPR018995 RNA_synth_NSP10_coronavirus
IPR037204 NSP7_sf
IPR014827 Viral_protease
IPR038123 NSP4_C_sf
IPR008740 Peptidase_C30
IPR014828 NSP7
IPR032505 Corona_NSP4_C
IPR014822 NSP9
IPR032592 NAR_dom
IPR002589 Macro_dom
IPR009003 Peptidase_S1_PA
IPR036499 NSP9_sf
IPR013016 Peptidase_C30/C16
IPR038400 Nsp3_coronavir_sf
IPR014829 NSP8
IPR036333 NSP10_sf
IPR042570 NAR_sf
IPR038083 R1a/1ab
SUPFAM
ProteinModelPortal
PDB
6NUS
E-value=0,
Score=3719
Ontologies
GO
GO:0016021 C:integral component of membrane
GO:0044220 C:host cell perinuclear region of cytoplasm
GO:0003723 F:RNA binding
GO:0008270 F:zinc ion binding
GO:0003968 F:RNA-directed 5'-3' RNA polymerase activity
GO:0039648 P:modulation by virus of host protein ubiquitination
GO:0039595 P:induction by virus of catabolism of host mRNA
GO:0019082 P:viral protein processing
GO:0004197 F:cysteine-type endopeptidase activity
GO:0039520 P:induction by virus of host autophagy
GO:0036459 F:thiol-dependent ubiquitinyl hydrolase activity
GO:0039548 P:suppression by virus of host IRF3 activity
GO:0008242 F:omega peptidase activity
GO:0033644 C:host cell membrane
GO:0019079 P:viral genome replication
GO:0039579 P:suppression by virus of host ISG15 activity
GO:0039502 P:suppression by virus of host type I interferon-mediated signaling pathway
GO:0044220 C:host cell perinuclear region of cytoplasm
GO:0003723 F:RNA binding
GO:0008270 F:zinc ion binding
GO:0003968 F:RNA-directed 5'-3' RNA polymerase activity
GO:0039648 P:modulation by virus of host protein ubiquitination
GO:0039595 P:induction by virus of catabolism of host mRNA
GO:0019082 P:viral protein processing
GO:0004197 F:cysteine-type endopeptidase activity
GO:0039520 P:induction by virus of host autophagy
GO:0036459 F:thiol-dependent ubiquitinyl hydrolase activity
GO:0039548 P:suppression by virus of host IRF3 activity
GO:0008242 F:omega peptidase activity
GO:0033644 C:host cell membrane
GO:0019079 P:viral genome replication
GO:0039579 P:suppression by virus of host ISG15 activity
GO:0039502 P:suppression by virus of host type I interferon-mediated signaling pathway
Subcellular Location
From MSLVP
Capsid
From Uniprot
Host membrane
Host cytoplasm
nsp7, nsp8, nsp9 and nsp10 are localized in cytoplasmic foci, largely perinuclear. Late in infection, they merge into confluent complexes (By similarity). With evidence from 5 publications.
Host cytoplasm
nsp7, nsp8, nsp9 and nsp10 are localized in cytoplasmic foci, largely perinuclear. Late in infection, they merge into confluent complexes (By similarity). With evidence from 5 publications.
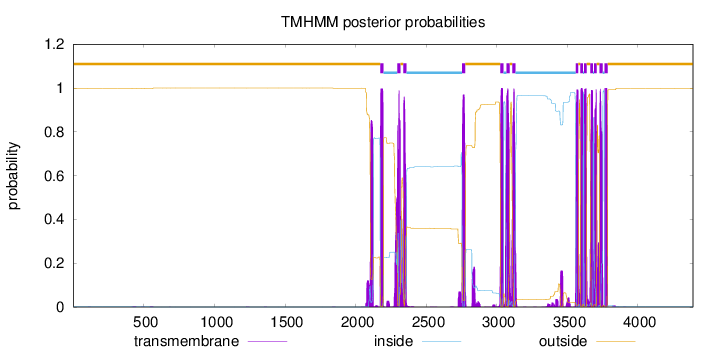
Topology
Length:
4391
Number of predicted TMHs:
14
Exp number of AAs in TMHs:
351.463199999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00122
outside
1 - 2174
TMhelix
2175 - 2197
inside
2198 - 2296
TMhelix
2297 - 2319
outside
2320 - 2338
TMhelix
2339 - 2361
inside
2362 - 2755
TMhelix
2756 - 2778
outside
2779 - 3026
TMhelix
3027 - 3049
inside
3050 - 3069
TMhelix
3070 - 3092
outside
3093 - 3111
TMhelix
3112 - 3134
inside
3135 - 3560
TMhelix
3561 - 3578
outside
3579 - 3592
TMhelix
3593 - 3612
inside
3613 - 3618
TMhelix
3619 - 3638
outside
3639 - 3663
TMhelix
3664 - 3683
inside
3684 - 3689
TMhelix
3690 - 3710
outside
3711 - 3729
TMhelix
3730 - 3752
inside
3753 - 3764
TMhelix
3765 - 3787
outside
3788 - 4391
Population Genetic Test Statistics
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 |
Protein Protein |
YP_009047202.1 YP_009047203.1 |
|
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 |
Protein Protein |
YP_007188577.3 YP_007188578.1 |
|
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 |
Protein |
YP_009361855.1 |
|
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.