Strain
USA_HS_2004_AY585228
(Region: USA; Strain: Human coronavirus OC43 strain ATCC VR-759, complete genome.; Date: 2004)
Gene
spike surface glycoprotein
Description
Annotated in NCBI,
spike surface glycoprotein
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGTTTTTGATACTTTTAATTTCCTTACCAACGGCTTTTGCTGTTATAGGAGATTTAAAGTGTACTTCAGATAATATTAATGATAAAGACACCGGTCCTCCTCCTATAAGTACTGATACTGTTGATGTTACTAATGGTTTGGGTACTTATTATGTTTTAGATCGTGTGTATTTAAATACTACGTTGTTTCTTAATGGTTATTACCCTACTTCAGGTTCCACATATCGTAATATGGCACTGAAGGGAAGTGTACTATTGAGCAGACTATGGTTTAAACCACCATTTCTTTCTGATTTTATTAATGGTATTTTTGCTAAGGTCAAAAATACCAAGGTTATTAAAGATCGTGTAATGTATAGTGAGTTCCCTGCTATAACTATAGGTAGTACTTTTGTAAATACATCCTATAGTGTGGTAGTACAACCACGTACAATCAATTCAACACAGGATGGTGATAATAAATTACAAGGTCTTTTAGAGGTCTCTGTTTGCCAGTATAATATGTGCGAGTACCCACAAACGATTTGTCATCCTAACCTGGGTAATCATCGCAAAGAACTATGGCATTTGGATACAGGTGTTGTTTCCTGTTTATATAAGCGTAATTTCACATATGATGTGAATGCTGATTATTTGTATTTTCATTTTTATCAAGAAGGTGGTACTTTTTATGCATATTTTACAGACACTGGTGTTGTTACTAAGTTTTTGTTTAATGTTTATTTAGGCATGGCGCTTTCACACTATTATGTCATGCCTCTGACTTGTAATAGTAAGCTTACTTTAGAATATTGGGTTACACCTCTCACTTCTAGACAATATTTACTCGCTTTCAATCAAGATGGTATTATTTTTAATGCTGTTGATTGTATGAGTGATTTTATGAGTGAGATTAAGTGTAAAACACAATCTATAGCACCACCTACTGGTGTTTATGAATTAAACGGTTACACTGTTCAGCCAATCGCAGATGTTTACCGACGTAAACCTAATCTTCCCAATTGCAATATAGAAGCTTGGCTTAATGATAAGTCGGTGCCCTCTCCATTAAATTGGGAACGTAAGACATTTTCAAATTGTAATTTTAATATGAGCAGCCTGATGTCTTTTATTCAGGCAGACTCATTTACTTGTAATAATATTGATGCTGCTAAGATATATGGTATGTGTTTTTCCAGCATAACTATAGATAAGTTTGCTATACCCAATGGCAGGAAGGTTGACCTACAATTGGGTAATTTGGGCTATTTGCAGTCATTTAACTATAGAATTGATACTACTGCAACAAGTTGTCAGTTGTATTATAATTTACCTGCTGCTAATGTTTCTGTTAGCAGGTTTAATCCTTCTACTTGGAATAAGAGATTTGGTTTTATAGAAGATTCTGTTTTTAAGCCTCGACCTGCAGGTGTTCTTACTAATCATGATGTAGTTTATGCACAACACTGTTTCAAAGCTCCTAAAAATTTCTGTCCGTGTAAATTGAATGGTTCGTGTGTAGGTAGTGGTCCTGGTAAAAATAATGGTATAGGCACTTGTCCTGCAGGTACTAATTATTTAACTTGTGATAATTTGTGCACTCCTGATCCTATTACATTTACAGGTACTTATAAGTGCCCCCAAACTAAATCTTTAGTTGGCATAGGTGAGCACTGTTCGGGTCTTGCTGTTAAAAGTGATTATTGTGGAGGCAATTCTTGTACTTGCCGACCACAAGCATTTTTGGGTTGGTCTGCAGACTCTTGTTTACAAGGAGACAAGTGTAATATTTTTGCTAATTTTATTTTGCATGATGTTAATAGTGGTCTTACTTGTTCTACTGATTTACAAAAAGCTAACACAGACATAATTCTTGGTGTTTGTGTTAATTATGACCTCTATGGTATTTTAGGCCAAGGCATTTTTGTTGAGGTTAATGCGACTTATTATAATAGTTGGCAGAACCTTTTATATGATTCTAATGGTAATCTCTACGGTTTTAGAGACTACATAACAAACAGAACTTTTATGATTCGTAGTTGCTATAGCGGTCGTGTTTCTGCGGCCTTTCACGCTAACTCTTCCGAACCAGCATTGCTATTTCGGAATATTAAATGCAACTACGTTTTTAATAATAGTCTTACACGACAGCTGCAACCCATTAACTATTTTGATAGTTATCTTGGTTGTGTTGTCAATGCTTATAATAGTACTGCTATTTCTGTTCAAACATGTGATCTCACAGTAGGTAGTGGTTACTGTGTGGATTACTCTAAAAACAGACGAAGTCGTGGAGCGATTACCACTGGTTATCGGTTTACTAATTTTGAGCCATTTACTGTTAATTCAGTAAACGATAGTTTAGAACCTGTAGGTGGTTTGTATGAAATTCAAATACCTTCAGAGTTTACTATAGGTAATATGGTGGAGTTTATTCAAACAAGCTCTCCTAAAGTTACTATTGATTGTGCTGCATTTGTCTGTGGTGATTATGCAGCATGTAAATCACAGTTGGTTGAATATGGTAGTTTCTGTGATAACATTAATGCCATACTCACAGAAGTAAATGAACTACTTGACACTACACAGTTGCAAGTAGCTAATAGTTTAATGAATGGTGTTACTCTTAGCACTAAGCTTAAAGATGGCGTTAATTTCAATGTAGACGACATCAATTTTTCCCCTGTATTAGGTTGTCTAGGCAGCGAATGTAGTAAAGCTTCCAGTAGATCTGCTATAGAGGATTTACTTTTTGATAAAGTAAAGTTATCTGATGTCGGTTTTGTTGAGGCTTATAATAATTGTACAGGAGGTGCCGAAATTAGGGACCTCATTTGTGTGCAAAGTTATAAAGGCATCAAAGTGTTGCCTCCACTGCTCTCAGAAAATCAGATCAGTGGATACACTTTGGCTGCCACCTCTGCTAGTCTATTTCCTCCTTGGACAGCAGCAGCAGGTGTACCATTTTATTTAAATGTTCAGTATCGCATTAATGGGCTTGGTGTCACCATGGATGTGCTAAGTCAAAATCAAAAGCTTATTGCTAATGCATTTAACAATGCCCTTTATGCTATTCAGGAAGGGTTCGATGCAACTAATTCTGCTTTAGTTAAAATTCAAGCTGTTGTTAATGCAAATGCTGAAGCTCTTAATAACTTATTGCAACAACTCTCTAATAGATTTGGTGCTATAAGTGCTTCTTTACAAGAAATTCTATCTAGACTTGATGCTCTTGAAGCGGAAGCTCAGATAGATAGACTTATTAATGGTCGTCTTACCGCTCTTAATGCTTATGTTTCTCAACAGCTTAGTGATTCTACACTGGTAAAATTTAGTGCAGCACAAGCTATGGAGAAGGTTAATGAATGTGTCAAAAGCCAATCATCTAGGATAAATTTCTGTGGTAATGGTAATCATATTATATCATTAGTGCAGAATGCTCCATATGGTTTGTATTTTATCCACTTTAGTTATGTCCCTACTAAGTATGTCACAGCGAGGGTTAGTCCTGGTCTGTGCATTGCTGGTGATAGAGGTATAGCTCCTAAGAGTGGTTATTTTGTTAATGTAAATAATACTTGGATGTACACTGGTAGTGGTTACTACTACCCTGAACCTATAACTGAAAATAATGTTGTTGTTATGAGTACCTGCGCTGTTAATTATACTAAAGCGCCGTATGTAATGCTGAACACTTCAATACCCAACCTTCCTGATTTTAAGGAAGAGTTGGATCAATGGTTTAAAAATCAAACATCAGTGGCACCAGATTTGTCACTTGATTATATAAATGTTACATTCTTGGACCTACAAGTTGAAATGAATAGGTTACAGGAGGCAATAAAAGTCTTAAATCAGAGCTACATCAATCTCAAGGACATTGGTACATATGAATATTATGTAAAATGGCCTTGGTATGTATGGCTTTTAATCTGCCTTGCTGGTGTAGCTATGCTTGTTTTACTATTCTTCATATGCTGTTGTACAGGATGTGGGACTAGTTGTTTTAAGAAATGTGGTGGTTGTTGTGATGATTATACTGGATACCAGGAGTTAGTAATCAAAACTTCACATGACGACTAA
Protein
MFLILLISLPTAFAVIGDLKCTSDNINDKDTGPPPISTDTVDVTNGLGTYYVLDRVYLNTTLFLNGYYPTSGSTYRNMALKGSVLLSRLWFKPPFLSDFINGIFAKVKNTKVIKDRVMYSEFPAITIGSTFVNTSYSVVVQPRTINSTQDGDNKLQGLLEVSVCQYNMCEYPQTICHPNLGNHRKELWHLDTGVVSCLYKRNFTYDVNADYLYFHFYQEGGTFYAYFTDTGVVTKFLFNVYLGMALSHYYVMPLTCNSKLTLEYWVTPLTSRQYLLAFNQDGIIFNAVDCMSDFMSEIKCKTQSIAPPTGVYELNGYTVQPIADVYRRKPNLPNCNIEAWLNDKSVPSPLNWERKTFSNCNFNMSSLMSFIQADSFTCNNIDAAKIYGMCFSSITIDKFAIPNGRKVDLQLGNLGYLQSFNYRIDTTATSCQLYYNLPAANVSVSRFNPSTWNKRFGFIEDSVFKPRPAGVLTNHDVVYAQHCFKAPKNFCPCKLNGSCVGSGPGKNNGIGTCPAGTNYLTCDNLCTPDPITFTGTYKCPQTKSLVGIGEHCSGLAVKSDYCGGNSCTCRPQAFLGWSADSCLQGDKCNIFANFILHDVNSGLTCSTDLQKANTDIILGVCVNYDLYGILGQGIFVEVNATYYNSWQNLLYDSNGNLYGFRDYITNRTFMIRSCYSGRVSAAFHANSSEPALLFRNIKCNYVFNNSLTRQLQPINYFDSYLGCVVNAYNSTAISVQTCDLTVGSGYCVDYSKNRRSRGAITTGYRFTNFEPFTVNSVNDSLEPVGGLYEIQIPSEFTIGNMVEFIQTSSPKVTIDCAAFVCGDYAACKSQLVEYGSFCDNINAILTEVNELLDTTQLQVANSLMNGVTLSTKLKDGVNFNVDDINFSPVLGCLGSECSKASSRSAIEDLLFDKVKLSDVGFVEAYNNCTGGAEIRDLICVQSYKGIKVLPPLLSENQISGYTLAATSASLFPPWTAAAGVPFYLNVQYRINGLGVTMDVLSQNQKLIANAFNNALYAIQEGFDATNSALVKIQAVVNANAEALNNLLQQLSNRFGAISASLQEILSRLDALEAEAQIDRLINGRLTALNAYVSQQLSDSTLVKFSAAQAMEKVNECVKSQSSRINFCGNGNHIISLVQNAPYGLYFIHFSYVPTKYVTARVSPGLCIAGDRGIAPKSGYFVNVNNTWMYTGSGYYYPEPITENNVVVMSTCAVNYTKAPYVMLNTSIPNLPDFKEELDQWFKNQTSVAPDLSLDYINVTFLDLQVEMNRLQEAIKVLNQSYINLKDIGTYEYYVKWPWYVWLLICLAGVAMLVLLFFICCCTGCGTSCFKKCGGCCDDYTGYQELVIKTSHDD
Summary
Function
Spike protein S1: attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Subunit
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes.
Similarity
Belongs to the betacoronaviruses spike protein family.
Keywords
Coiled coil
Disulfide bond
Fusion of virus membrane with host endosomal membrane
Fusion of virus membrane with host membrane
Glycoprotein
Host cell membrane
Host membrane
Host-virus interaction
Lipoprotein
Membrane
Palmitate
Reference proteome
Signal
Transmembrane
Transmembrane helix
Viral attachment to host cell
Viral envelope protein
Viral penetration into host cytoplasm
Virion
Virulence
Virus entry into host cell
Feature
chain Spike protein S2
Uniprot
Q696P8
P36334
A0A0G2RC85
U3M7P4
U3M7N2
U3M6X6
+ More
U3M6B4 A0A2D1QW75 A0A0G2RC97 U3M6B6 U3M712 U3M6B2 U3M6X0 U3M716 U3M6V9 A0A385H6Q3 Q4VIF0 A0A385H6H8 U3M7L8 U3M7L4 A0A0G2RCC7 A0A0G2RD21 Q4VIE4 A0A0G2RC89 A0A0G2RCA9 A0A385H6Q9 A0A482KC70 A0A385H6T1 A0A5B9MWF9 A0A3S6FIX9 A0A385H5W2 A0A0B4N6Y8 A0A3S6FJG2 A0A0B4Q7Z0 A0A0B4UE97 A0A5B9MRW3 A0A0B4N6J3 A0A1W6DZX7 A0A0B4N7I3 A0A2R3TWV1 A0A0G2RD25 A0A482KB25 A0A0K0L9B0 A0A0G2RCA0 A0A0B4N6Y5 A0A0B4N5X0 A0A0B4Q9M3 A0A0B4N6J6 A0A0B4N687 U3M6Y8 A0A0G2RC93 A0A513ZT20 U3M7P6 A0A0B4Q908 U3M7M6 A0A224AU30 U3M7N6 U3M7N4 U3M7L6 U3M6Z7 U3M6W6 U3M7Q0 U3M7L0 U3M704 U3M7Q6 U3M7Q4 U3M7M0 Q4VID5 U3M701 Q4VIE8 A0A0G2RD16 G5CJB9 G5CJB1 A0A140E065 A0A0B4N5X3 A0A0K0KPM4 A0A0G2RCA7 A0A0B4Q8M2 A0A0B4N6J8 A0A0K0L8I8 U3M6Z3 A0A0A0YL58 A0A0B4N7J1 A0A0A0YKA3 A0A0B4N6A9 A0A0B4N6B2 A0A1C8Z4S1 A0A1S5VGH2 A0A0B4N6K6 A0A0B4N6B8 A0A0B4N6K8 A0A0B4N6B6 A0A0B4N708 A0A0B4N716 A0A386BY51 A0A0B4N7J5 A0A0B4N6A6 A0A0K0KPR8 A0A0B4N711 A0A0B4N5Z6
U3M6B4 A0A2D1QW75 A0A0G2RC97 U3M6B6 U3M712 U3M6B2 U3M6X0 U3M716 U3M6V9 A0A385H6Q3 Q4VIF0 A0A385H6H8 U3M7L8 U3M7L4 A0A0G2RCC7 A0A0G2RD21 Q4VIE4 A0A0G2RC89 A0A0G2RCA9 A0A385H6Q9 A0A482KC70 A0A385H6T1 A0A5B9MWF9 A0A3S6FIX9 A0A385H5W2 A0A0B4N6Y8 A0A3S6FJG2 A0A0B4Q7Z0 A0A0B4UE97 A0A5B9MRW3 A0A0B4N6J3 A0A1W6DZX7 A0A0B4N7I3 A0A2R3TWV1 A0A0G2RD25 A0A482KB25 A0A0K0L9B0 A0A0G2RCA0 A0A0B4N6Y5 A0A0B4N5X0 A0A0B4Q9M3 A0A0B4N6J6 A0A0B4N687 U3M6Y8 A0A0G2RC93 A0A513ZT20 U3M7P6 A0A0B4Q908 U3M7M6 A0A224AU30 U3M7N6 U3M7N4 U3M7L6 U3M6Z7 U3M6W6 U3M7Q0 U3M7L0 U3M704 U3M7Q6 U3M7Q4 U3M7M0 Q4VID5 U3M701 Q4VIE8 A0A0G2RD16 G5CJB9 G5CJB1 A0A140E065 A0A0B4N5X3 A0A0K0KPM4 A0A0G2RCA7 A0A0B4Q8M2 A0A0B4N6J8 A0A0K0L8I8 U3M6Z3 A0A0A0YL58 A0A0B4N7J1 A0A0A0YKA3 A0A0B4N6A9 A0A0B4N6B2 A0A1C8Z4S1 A0A1S5VGH2 A0A0B4N6K6 A0A0B4N6B8 A0A0B4N6K8 A0A0B4N6B6 A0A0B4N708 A0A0B4N716 A0A386BY51 A0A0B4N7J5 A0A0B4N6A6 A0A0K0KPR8 A0A0B4N711 A0A0B4N5Z6
Pubmed
EMBL
AY585229
AAT84362.1
L14643
S62886
Z21849
Z32768
+ More
Z32769 AY585228 AY391777 KF963229 AIX10748.1 KF530093 AGT51740.1 KF530087 AGT51680.1 KF530065 KF530066 KF530085 AGT51461.1 AGT51471.1 AGT51660.1 KF530060 KF530077 KF530083 KF530086 AGT51412.1 AGT51580.1 AGT51640.1 AGT51670.1 KY014281 ATP16757.1 KF963233 AIX10752.1 KF530061 AGT51422.1 KF530073 AGT51541.1 KF530059 KF530075 AGT51402.1 AGT51561.1 KF530064 KF530072 KF530092 AGT51451.1 AGT51531.1 AGT51730.1 KF530074 KF530090 KF530097 AGT51551.1 AGT51710.1 AGT51780.1 KF530062 AGT51431.1 KU745540 KU745541 MG197709 MG197719 ANZ78841.1 ANZ78842.1 AXX83297.1 AXX83357.1 AY903454 AY903456 AY903457 AAX84791.1 AAX84793.1 AAX84794.1 MG197718 AXX83351.1 KF530080 AGT51610.1 KF530078 AGT51590.1 KF963230 AIX10749.1 KF963236 AIX10755.1 AY903459 AAX85669.1 KF963234 AIX10753.1 KF963237 AIX10756.1 MG197711 MG197720 AXX83309.1 AXX83363.1 MK303625 QBQ01839.1 MG197714 AXX83327.1 MN310476 QEG03803.1 KU745546 ANZ78847.1 MG197721 AXX83369.1 KF572813 KF923898 AIL49493.1 AIV41849.1 KU745542 ANZ78843.1 KF572810 KF923888 AIL49490.1 AIV41789.1 KF572809 KP198610 AIL49489.1 AJC98127.1 MN306043 QEG03776.1 KF572806 AIL49486.1 KY967360 ARK08670.1 KF572804 KP198611 AIL49484.1 AJC98135.1 MF314143 AVQ05264.1 KF963241 AIX10760.1 MK327281 QBP84759.1 KF923895 AIV41831.1 KF963238 KY014282 AIX10757.1 ATP16767.1 KF572808 KF923887 AIL49488.1 AIV41783.1 KF572805 KF923886 AIL49485.1 AIV41777.1 KF572814 KF923906 AIL49494.1 AIV41897.1 KF572811 AIL49491.1 KF572807 KF923889 AIL49487.1 AIV41795.1 KF530067 AGT51481.1 KF963239 AIX10758.1 MN026165 QDH43726.1 KF530082 KF530094 AGT51630.1 AGT51750.1 KF572812 KF923896 AIL49492.1 AIV41837.1 KF530084 AGT51650.1 LC315649 BBA20982.1 KF530089 AGT51700.1 KF530088 KF530091 AGT51690.1 AGT51720.1 KF530079 AGT51600.1 KF530069 AGT51501.1 KF530063 AGT51441.1 KF530095 KF530096 AGT51760.1 AGT51770.1 KF530076 AGT51570.1 KF530071 AGT51521.1 KF530099 AGT51800.1 KF530098 AGT51790.1 KF530081 AGT51620.1 AY903455 AY903460 KF963235 AAX84792.1 AAX85678.1 AIX10754.1 KF530070 AGT51511.1 AY903458 AAX84795.1 KF963231 AIX10750.1 JN129835 AEN19366.1 JN129834 AEN19358.1 KU131570 AMK59677.1 KF572816 AIL49496.1 KF572840 AIL49520.1 KF963232 AIX10751.1 KF572818 KF923900 AIL49498.1 AIV41861.1 KF572817 KF923899 AIL49497.1 AIV41855.1 KF923905 AIV41891.1 KF530068 AGT51491.1 KJ958218 AIX09799.1 KF572825 KF923925 AIL49505.1 AIV42011.1 KJ958219 AIX09807.1 KF572846 KF572850 KF572857 KF923892 KF923909 KF923915 AIL49526.1 AIL49530.1 AIL49537.1 AIV41813.1 AIV41915.1 AIV41951.1 KF572819 KF572842 KF572844 KF572847 KF572852 KF572853 KF572854 KF572855 KF572860 KF572861 KF923890 KF923891 KF923894 KF923901 KF923911 KF923912 KF923913 KF923919 KF923920 AIL49499.1 AIL49522.1 AIL49524.1 AIL49527.1 AIL49532.1 AIL49533.1 AIL49534.1 AIL49535.1 AIL49540.1 AIL49541.1 AIV41801.1 AIV41807.1 AIV41825.1 AIV41867.1 AIV41927.1 AIV41933.1 AIV41939.1 AIV41975.1 AIV41981.1 KX344031 AOL02453.1 KX538970 KX538972 KX538976 AQN78704.1 AQN78720.1 AQN78752.1 KF572833 KF923904 AIL49513.1 AIV41885.1 KF572821 KF572824 KF572826 KF572837 KF572867 KF572869 KF572871 AIL49501.1 AIL49504.1 AIL49506.1 AIL49517.1 AIL49547.1 AIL49549.1 AIL49551.1 KF572838 AIL49518.1 KF572865 AIL49545.1 KF572851 KF923910 AIL49531.1 AIV41921.1 KF572866 KF923922 AIL49546.1 AIV41993.1 MH685718 AYC76638.1 KF572836 AIL49516.1 KF572845 AIL49525.1 KF572829 AIL49509.1 KF572856 KF923914 AIL49536.1 AIV41945.1 KF572848 KF923907 AIL49528.1 AIV41903.1
Z32769 AY585228 AY391777 KF963229 AIX10748.1 KF530093 AGT51740.1 KF530087 AGT51680.1 KF530065 KF530066 KF530085 AGT51461.1 AGT51471.1 AGT51660.1 KF530060 KF530077 KF530083 KF530086 AGT51412.1 AGT51580.1 AGT51640.1 AGT51670.1 KY014281 ATP16757.1 KF963233 AIX10752.1 KF530061 AGT51422.1 KF530073 AGT51541.1 KF530059 KF530075 AGT51402.1 AGT51561.1 KF530064 KF530072 KF530092 AGT51451.1 AGT51531.1 AGT51730.1 KF530074 KF530090 KF530097 AGT51551.1 AGT51710.1 AGT51780.1 KF530062 AGT51431.1 KU745540 KU745541 MG197709 MG197719 ANZ78841.1 ANZ78842.1 AXX83297.1 AXX83357.1 AY903454 AY903456 AY903457 AAX84791.1 AAX84793.1 AAX84794.1 MG197718 AXX83351.1 KF530080 AGT51610.1 KF530078 AGT51590.1 KF963230 AIX10749.1 KF963236 AIX10755.1 AY903459 AAX85669.1 KF963234 AIX10753.1 KF963237 AIX10756.1 MG197711 MG197720 AXX83309.1 AXX83363.1 MK303625 QBQ01839.1 MG197714 AXX83327.1 MN310476 QEG03803.1 KU745546 ANZ78847.1 MG197721 AXX83369.1 KF572813 KF923898 AIL49493.1 AIV41849.1 KU745542 ANZ78843.1 KF572810 KF923888 AIL49490.1 AIV41789.1 KF572809 KP198610 AIL49489.1 AJC98127.1 MN306043 QEG03776.1 KF572806 AIL49486.1 KY967360 ARK08670.1 KF572804 KP198611 AIL49484.1 AJC98135.1 MF314143 AVQ05264.1 KF963241 AIX10760.1 MK327281 QBP84759.1 KF923895 AIV41831.1 KF963238 KY014282 AIX10757.1 ATP16767.1 KF572808 KF923887 AIL49488.1 AIV41783.1 KF572805 KF923886 AIL49485.1 AIV41777.1 KF572814 KF923906 AIL49494.1 AIV41897.1 KF572811 AIL49491.1 KF572807 KF923889 AIL49487.1 AIV41795.1 KF530067 AGT51481.1 KF963239 AIX10758.1 MN026165 QDH43726.1 KF530082 KF530094 AGT51630.1 AGT51750.1 KF572812 KF923896 AIL49492.1 AIV41837.1 KF530084 AGT51650.1 LC315649 BBA20982.1 KF530089 AGT51700.1 KF530088 KF530091 AGT51690.1 AGT51720.1 KF530079 AGT51600.1 KF530069 AGT51501.1 KF530063 AGT51441.1 KF530095 KF530096 AGT51760.1 AGT51770.1 KF530076 AGT51570.1 KF530071 AGT51521.1 KF530099 AGT51800.1 KF530098 AGT51790.1 KF530081 AGT51620.1 AY903455 AY903460 KF963235 AAX84792.1 AAX85678.1 AIX10754.1 KF530070 AGT51511.1 AY903458 AAX84795.1 KF963231 AIX10750.1 JN129835 AEN19366.1 JN129834 AEN19358.1 KU131570 AMK59677.1 KF572816 AIL49496.1 KF572840 AIL49520.1 KF963232 AIX10751.1 KF572818 KF923900 AIL49498.1 AIV41861.1 KF572817 KF923899 AIL49497.1 AIV41855.1 KF923905 AIV41891.1 KF530068 AGT51491.1 KJ958218 AIX09799.1 KF572825 KF923925 AIL49505.1 AIV42011.1 KJ958219 AIX09807.1 KF572846 KF572850 KF572857 KF923892 KF923909 KF923915 AIL49526.1 AIL49530.1 AIL49537.1 AIV41813.1 AIV41915.1 AIV41951.1 KF572819 KF572842 KF572844 KF572847 KF572852 KF572853 KF572854 KF572855 KF572860 KF572861 KF923890 KF923891 KF923894 KF923901 KF923911 KF923912 KF923913 KF923919 KF923920 AIL49499.1 AIL49522.1 AIL49524.1 AIL49527.1 AIL49532.1 AIL49533.1 AIL49534.1 AIL49535.1 AIL49540.1 AIL49541.1 AIV41801.1 AIV41807.1 AIV41825.1 AIV41867.1 AIV41927.1 AIV41933.1 AIV41939.1 AIV41975.1 AIV41981.1 KX344031 AOL02453.1 KX538970 KX538972 KX538976 AQN78704.1 AQN78720.1 AQN78752.1 KF572833 KF923904 AIL49513.1 AIV41885.1 KF572821 KF572824 KF572826 KF572837 KF572867 KF572869 KF572871 AIL49501.1 AIL49504.1 AIL49506.1 AIL49517.1 AIL49547.1 AIL49549.1 AIL49551.1 KF572838 AIL49518.1 KF572865 AIL49545.1 KF572851 KF923910 AIL49531.1 AIV41921.1 KF572866 KF923922 AIL49546.1 AIV41993.1 MH685718 AYC76638.1 KF572836 AIL49516.1 KF572845 AIL49525.1 KF572829 AIL49509.1 KF572856 KF923914 AIL49536.1 AIV41945.1 KF572848 KF923907 AIL49528.1 AIV41903.1
Proteomes
UP000100580
UP000007552
UP000180344
UP000105266
UP000106834
UP000147052
+ More
UP000165173 UP000096360 UP000136788 UP000159503 UP000167348 UP000168172 UP000143333 UP000149702 UP000115705 UP000126877 UP000154668 UP000147585 UP000149594 UP000170317 UP000100971 UP000165185 UP000161137 UP000142982 UP000104994 UP000134871 UP000133796 UP000161839 UP000143341 UP000114688 UP000173306 UP000137844 UP000122365 UP000104124 UP000114034 UP000174077 UP000100004 UP000109543 UP000150595 UP000160021 UP000143194 UP000155676 UP000172586 UP000140040 UP000158566 UP000119922 UP000097333 UP000109321 UP000168290 UP000140401 UP000159995 UP000117387 UP000130586 UP000173822 UP000147591 UP000121854 UP000119882 UP000128148 UP000119762 UP000126267 UP000173555 UP000099221 UP000130040 UP000142623 UP000166197 UP000107048 UP000115819 UP000116223 UP000119058 UP000119993 UP000123825 UP000132146 UP000136628 UP000137532 UP000165756 UP000136483 UP000114169 UP000117554 UP000115343
UP000165173 UP000096360 UP000136788 UP000159503 UP000167348 UP000168172 UP000143333 UP000149702 UP000115705 UP000126877 UP000154668 UP000147585 UP000149594 UP000170317 UP000100971 UP000165185 UP000161137 UP000142982 UP000104994 UP000134871 UP000133796 UP000161839 UP000143341 UP000114688 UP000173306 UP000137844 UP000122365 UP000104124 UP000114034 UP000174077 UP000100004 UP000109543 UP000150595 UP000160021 UP000143194 UP000155676 UP000172586 UP000140040 UP000158566 UP000119922 UP000097333 UP000109321 UP000168290 UP000140401 UP000159995 UP000117387 UP000130586 UP000173822 UP000147591 UP000121854 UP000119882 UP000128148 UP000119762 UP000126267 UP000173555 UP000099221 UP000130040 UP000142623 UP000166197 UP000107048 UP000115819 UP000116223 UP000119058 UP000119993 UP000123825 UP000132146 UP000136628 UP000137532 UP000165756 UP000136483 UP000114169 UP000117554 UP000115343
PRIDE
Interpro
SUPFAM
SSF143587
SSF143587
Gene 3D
ProteinModelPortal
Q696P8
P36334
A0A0G2RC85
U3M7P4
U3M7N2
U3M6X6
+ More
U3M6B4 A0A2D1QW75 A0A0G2RC97 U3M6B6 U3M712 U3M6B2 U3M6X0 U3M716 U3M6V9 A0A385H6Q3 Q4VIF0 A0A385H6H8 U3M7L8 U3M7L4 A0A0G2RCC7 A0A0G2RD21 Q4VIE4 A0A0G2RC89 A0A0G2RCA9 A0A385H6Q9 A0A482KC70 A0A385H6T1 A0A5B9MWF9 A0A3S6FIX9 A0A385H5W2 A0A0B4N6Y8 A0A3S6FJG2 A0A0B4Q7Z0 A0A0B4UE97 A0A5B9MRW3 A0A0B4N6J3 A0A1W6DZX7 A0A0B4N7I3 A0A2R3TWV1 A0A0G2RD25 A0A482KB25 A0A0K0L9B0 A0A0G2RCA0 A0A0B4N6Y5 A0A0B4N5X0 A0A0B4Q9M3 A0A0B4N6J6 A0A0B4N687 U3M6Y8 A0A0G2RC93 A0A513ZT20 U3M7P6 A0A0B4Q908 U3M7M6 A0A224AU30 U3M7N6 U3M7N4 U3M7L6 U3M6Z7 U3M6W6 U3M7Q0 U3M7L0 U3M704 U3M7Q6 U3M7Q4 U3M7M0 Q4VID5 U3M701 Q4VIE8 A0A0G2RD16 G5CJB9 G5CJB1 A0A140E065 A0A0B4N5X3 A0A0K0KPM4 A0A0G2RCA7 A0A0B4Q8M2 A0A0B4N6J8 A0A0K0L8I8 U3M6Z3 A0A0A0YL58 A0A0B4N7J1 A0A0A0YKA3 A0A0B4N6A9 A0A0B4N6B2 A0A1C8Z4S1 A0A1S5VGH2 A0A0B4N6K6 A0A0B4N6B8 A0A0B4N6K8 A0A0B4N6B6 A0A0B4N708 A0A0B4N716 A0A386BY51 A0A0B4N7J5 A0A0B4N6A6 A0A0K0KPR8 A0A0B4N711 A0A0B4N5Z6
U3M6B4 A0A2D1QW75 A0A0G2RC97 U3M6B6 U3M712 U3M6B2 U3M6X0 U3M716 U3M6V9 A0A385H6Q3 Q4VIF0 A0A385H6H8 U3M7L8 U3M7L4 A0A0G2RCC7 A0A0G2RD21 Q4VIE4 A0A0G2RC89 A0A0G2RCA9 A0A385H6Q9 A0A482KC70 A0A385H6T1 A0A5B9MWF9 A0A3S6FIX9 A0A385H5W2 A0A0B4N6Y8 A0A3S6FJG2 A0A0B4Q7Z0 A0A0B4UE97 A0A5B9MRW3 A0A0B4N6J3 A0A1W6DZX7 A0A0B4N7I3 A0A2R3TWV1 A0A0G2RD25 A0A482KB25 A0A0K0L9B0 A0A0G2RCA0 A0A0B4N6Y5 A0A0B4N5X0 A0A0B4Q9M3 A0A0B4N6J6 A0A0B4N687 U3M6Y8 A0A0G2RC93 A0A513ZT20 U3M7P6 A0A0B4Q908 U3M7M6 A0A224AU30 U3M7N6 U3M7N4 U3M7L6 U3M6Z7 U3M6W6 U3M7Q0 U3M7L0 U3M704 U3M7Q6 U3M7Q4 U3M7M0 Q4VID5 U3M701 Q4VIE8 A0A0G2RD16 G5CJB9 G5CJB1 A0A140E065 A0A0B4N5X3 A0A0K0KPM4 A0A0G2RCA7 A0A0B4Q8M2 A0A0B4N6J8 A0A0K0L8I8 U3M6Z3 A0A0A0YL58 A0A0B4N7J1 A0A0A0YKA3 A0A0B4N6A9 A0A0B4N6B2 A0A1C8Z4S1 A0A1S5VGH2 A0A0B4N6K6 A0A0B4N6B8 A0A0B4N6K8 A0A0B4N6B6 A0A0B4N708 A0A0B4N716 A0A386BY51 A0A0B4N7J5 A0A0B4N6A6 A0A0K0KPR8 A0A0B4N711 A0A0B4N5Z6
PDB
6OHW
E-value=0,
Score=5808
Ontologies
GO
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0016021 C:integral component of membrane
GO:0019031 C:viral envelope
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0020002 C:host cell plasma membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0046789 F:host cell surface receptor binding
GO:0030246 F:carbohydrate binding
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0016021 C:integral component of membrane
GO:0019031 C:viral envelope
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0020002 C:host cell plasma membrane
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0046789 F:host cell surface receptor binding
GO:0030246 F:carbohydrate binding
Subcellular Location
From MSLVP
Host nucleus > host nucleolus. Host cytoplasm. Secreted.
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 3 publications.
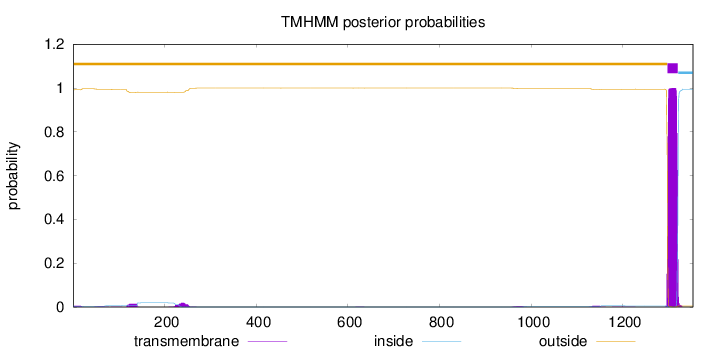
Topology
Length:
1353
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.04016
Exp number, first 60 AAs:
0.13052
Total prob of N-in:
0.00791
outside
1 - 1297
TMhelix
1298 - 1320
inside
1321 - 1353
Population Genetic Test Statistics
Pi
0.5695559
Theta
0.6939842
Tajima's D
-0.7732285
CLR
12.685631
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 |
Protein |
YP_005454245.1 |
|
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 |
Protein |
AAT84354.1 |
|
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 |
Protein |
NP_150077.1 |
|
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.