Strain
GERMANY_ERINACEINAE_2012_NC_022643
(Region: Germany; Strain: Betacoronavirus Erinaceus/VMC/DEU/2012 isolate ErinaceusCoV/2012-216/GER/2012, complete genome.; Date: 2012)
Gene
S
Description
Annotated in NCBI,
S
Location
GenBank Accession
Full name
Spike glycoprotein
Alternative Name
E2
Peplomer protein
Peplomer protein
Sequence
CDS
ATGATACGCTCAGCGTGTCTACTGATGTGCTTGTTAATGTTTATAAAAGCAACCCCAAGCGAGGGAACATGTATATCTGTTGACATGCAGCCCTCATATTTTATCAAAAATTGGTCTATGCCAATTAATATGAGTAAAGCAGATGGTGTTTTATACCCTGTTGGGCGTACTTATTCTAATATCACAATAACGCTTGAAGGGTTATTTCCACACCAAGCAGATCGTGGTGATATGTATGTTTACTCACAATCTCATAAAGGCAGGCCTTTCATTTCTAATTATTCTCTCGTTACTAATGATTTTGGTAATGGCATAGTAATTAGAATAGGTTCTGCTGCTAATAAAACTGGTAGTCCTATTATTTCTAATATTGCTAGTAACTTTCAGATTATGAAGAAACTTTATCCAGCTTTGCTCTTTGGTAGTGCTGTTGGTAAATTTCCTGCTAATAATAAAACAGGTGCTTATTTTAATCATAGCCTCGTTATTTTGCCCGAAAAATGTGGCACTGTTATTACAGCACTTTATTGTGTTTTAGAGCCCAAGAATGATACCAATTGTACTGGTAGTTCTGGTTATGTTAGTTATGTGCTGTTTGAAAATTTTAGTTGTGATGCAACTACTGATGGTAACTTAAAAAATAGTTTACAGCAGTGGTTTAATATTAAGGATTGTCTTTTTGAGAAAAATTACAATGTTACTGATGATGAGAGGGAAGAGTGGTTTGGAATTATCCAAGATCAGCAAGGTGTCCACCTCTATACATCTCGGAAGAATGGATATTCAAACAATATGTTTTTGTTTGCTACTTTACCAATTTATGATCAGATACTATATTATACTGTTATGCCGCGCAGTATTAATGCTAGTAATTATGCATCATATCATGCTTTTTCTGCATTTTATATTTACAAGTTACACAAAATCAACTATATGGTTGACTTTGATGTAAATGGTTATATCACAAGGGCTGTCGATTGTGGTTATAATGATTATACACAATTGCAATGCTCTTATGGTCAATTTGATATGGATTCTGGTGTTTATTCAGCTTCGTATTTTAATGCCCGCTCTAGAGGTTATTACTATGAAGCTGCAGAACTCACAGAATGTGACTTAGATGTATTGTTTAAAAACGATGCTCCTATAATAGCTAATTACAGTAGGCGAGTTTTTACTAATTGTAATTATAATTTAACAAAACTCCTTTCTCTTGTGCAAGTAGATGAGTTTGTTTGTCATAAAACAACACCAGAAGCGCTTGCTACTGGCTGTTATTCCTCTCTAACGGTAGACTGGTTTGCGCTGCCTTTTAGTATGAAGTCAACATTAGCTATAGGATCAGCAGAAGCTATATCTATGTTTAATTATAATCAGGATTATTCCAATCCTACGTGTAGAATTCATGCAGCTGTAACAGCTAATGTTTCAACAGCGTTGAATTTTACTGCTAATGCTAATTATGCTTATATTTCCAGGTGCCAAGGTGTTGATGGTAAACCAATTCTATTACAACCAGGACAAATGACTAATATTGCTTGTCGTTCTGGTGTTTTGGCACGGCCATCAGATGCAGATTATTTTGGCTATTCATTTCAAGGACGCAACTATTATTTAGGGCGTAAATCTTATAAACCTAAAACCGATGAGGGCGACGTTCAAATGGTTTATGTAATAACGCCTAAATATGATAAAGGCCCTGATACTGTATGCCCTCTTAAAGATATGGGTGCTGCCTCTTCTAGTTTAGATGGCTTGTTAGGGCAGTGCATAGATTATGACATTCATGGAGTAGTAGGCCGTGGTGTTTTTCAAGTTTGCAATACCACAGGAATTGCTAGTCAAATTTTTGTCTATGACGGATTTGGTAATATCATAGGCTTTCATTCTAAAAATGGTACATACTATTGTATGGCACCATGCGTATCAGTACCTGTTTCTGTAATTTATGATAAAAGCAGTGATGTTCATGCTACGCTTTATAGTAGCGTTGAGTGCAATCATATAAAATCAGTTGCAACAGTTTTTAGTCGTCAAACTGAGTCTAAGCTTAGATCGAGTGATAACGGCCTTTTACAAACAGCTGTAGGCTGTGTTATTGGATTCCGTAACACAAGTGACACTGTTGAAGATTGTACCTTACCTTTAGGCCAATCTCTCTGTGCAAAACCACCTTCTTTTTCCAGTCGTAGTACCAATGTTAATAACACATTTGCTTTAGTGCATATGTTGTTCCAAAATCCATTAAAAGTTAGTGTTCTCAATTCAACTGAGTTTCAGGTTTCAATACCTCAGAATTTTAGTTTTGGTATTACTGAAGAATTTATAGAAACATCCATACAAAAGGTTACTGTTGATTGTAAGCAGTATGTTTGTAATGGATTTGAGCGTTGTGAACAATTGTTGGTGCAGTATGGCCAATTTTGTTCAAAAATTAATCAAGCATTACATGGTGTTAATTTGCGCCAAGATGATGCTACTAAGAGTCTTTTTGAGGACATTAAAGTACCCCAATCAGTACCCTTAATGACAAGCCTCACTGGTGATTATAATTTATCATTATTCGAGGCACCTAATATTGGCAGTGGTAATAATAACTACCGCAGTGCTTTAGAAGATCTGCTTTTTGATAAGGTTACATTATCAGATCCAGGTTATATGAAAGGATATGACGAGTGCATGAAAAAAGGACCACCTAGTGCTAGGGACCTAATTTGTGCACAATATGTATCTGGTTATAAAGTTTTACCGCCTTTGTATGATGCTAATATGGAGGCTATGTATACTGCTTCTTTAACTGGTAGTATAGCAGGTTCTTTTTGGACAGGAGGACTATCGTCTGCTGCTGCACTACCATTTGCACAGTCAATGTTTTATCGTATGAATGGTATTGGCATCACTCAAAATGTTCTTATGAAGAACCAGAGGGAGATAGCCAATAAGTTCAACCAGGCATTAGGTGCTATGCAAACAGGATTCACAGCTACCAATCAAGCTTTTCAGAAGGTTCAAGATGTTGTGAATGCCAATGCACAAGCATTATCTAAATTAGCTAGTGAATTAGCTAACACATTCGGTGCTATTTCCGCTTCTATAGGTGACATCTTAAAACGTTTGGATGTTTTAGAACAGGAAGTGCAAATAGACCGCCTTATTAACGGACGTCTTACATCGTTGAATGCGTTTGTAAGTCAGCAACTGGTGAGGAGTGAAGCTGCAGCTCGCTCATCTCAGTTGGCTAAAGAAAAAATTAATGAATGTGTCAAAGCTCAATCTACACGCAGTGGGTTTTGCGGACAAGGCACTCATATAGTGTCATTTGTTATTAATGCCCCTAATGGATTTTATTTTATACATGTGGGTTATCACCCACAGGATTATGTAAATCAAACAGCTGCTTATGGCCTGTGTGATTCTTCTTCTAAGAAATGTATTGCAGCCAAGAATGGCTATTTTGTTAAAAATGATTCTGATAGTGGTAGTTGGTCTTATACAGGCAGCAGTTTTTATCAGCCTGAACCTATCACCACTTTTAACTCAAGGTTTGTTGAACCTGAGGTAACTTTTCAAAATTTAAGCAACAACCTTCCACCTCCACTCTTAAGTAATAACACTGATATTACTTTTGAGGATGAGCTGGAAGAGTTTTATAAAAATATTACGTCTGAGATACCTAATTTTGGTTCTCTCAGTCAGATTAATACCACTATGCTTAACTTGTCTCAAGAAATGAGCACTTTACAGCAAGTGGTCAAAGATTTAAACAACTCTTATATAGAGCTTAAAGAATTAGGCAATCACACATACTACCAAAAATGGCCGTGGTATGTGTGGTTGGGTTTCATTGCTGGTCTTTTGGCACTAGCATTATGTTTATTTTTCATACTGTGTTGCACAGGGTGTGGCACTAGTTGTCTAGGAAAAATAAGTTGTACTCGTTGTTGTGATAAGTATGATGATTTTGAAATGCCGGACAAAGTTCACATACACTAA
Protein
MIRSACLLMCLLMFIKATPSEGTCISVDMQPSYFIKNWSMPINMSKADGVLYPVGRTYSNITITLEGLFPHQADRGDMYVYSQSHKGRPFISNYSLVTNDFGNGIVIRIGSAANKTGSPIISNIASNFQIMKKLYPALLFGSAVGKFPANNKTGAYFNHSLVILPEKCGTVITALYCVLEPKNDTNCTGSSGYVSYVLFENFSCDATTDGNLKNSLQQWFNIKDCLFEKNYNVTDDEREEWFGIIQDQQGVHLYTSRKNGYSNNMFLFATLPIYDQILYYTVMPRSINASNYASYHAFSAFYIYKLHKINYMVDFDVNGYITRAVDCGYNDYTQLQCSYGQFDMDSGVYSASYFNARSRGYYYEAAELTECDLDVLFKNDAPIIANYSRRVFTNCNYNLTKLLSLVQVDEFVCHKTTPEALATGCYSSLTVDWFALPFSMKSTLAIGSAEAISMFNYNQDYSNPTCRIHAAVTANVSTALNFTANANYAYISRCQGVDGKPILLQPGQMTNIACRSGVLARPSDADYFGYSFQGRNYYLGRKSYKPKTDEGDVQMVYVITPKYDKGPDTVCPLKDMGAASSSLDGLLGQCIDYDIHGVVGRGVFQVCNTTGIASQIFVYDGFGNIIGFHSKNGTYYCMAPCVSVPVSVIYDKSSDVHATLYSSVECNHIKSVATVFSRQTESKLRSSDNGLLQTAVGCVIGFRNTSDTVEDCTLPLGQSLCAKPPSFSSRSTNVNNTFALVHMLFQNPLKVSVLNSTEFQVSIPQNFSFGITEEFIETSIQKVTVDCKQYVCNGFERCEQLLVQYGQFCSKINQALHGVNLRQDDATKSLFEDIKVPQSVPLMTSLTGDYNLSLFEAPNIGSGNNNYRSALEDLLFDKVTLSDPGYMKGYDECMKKGPPSARDLICAQYVSGYKVLPPLYDANMEAMYTASLTGSIAGSFWTGGLSSAAALPFAQSMFYRMNGIGITQNVLMKNQREIANKFNQALGAMQTGFTATNQAFQKVQDVVNANAQALSKLASELANTFGAISASIGDILKRLDVLEQEVQIDRLINGRLTSLNAFVSQQLVRSEAAARSSQLAKEKINECVKAQSTRSGFCGQGTHIVSFVINAPNGFYFIHVGYHPQDYVNQTAAYGLCDSSSKKCIAAKNGYFVKNDSDSGSWSYTGSSFYQPEPITTFNSRFVEPEVTFQNLSNNLPPPLLSNNTDITFEDELEEFYKNITSEIPNFGSLSQINTTMLNLSQEMSTLQQVVKDLNNSYIELKELGNHTYYQKWPWYVWLGFIAGLLALALCLFFILCCTGCGTSCLGKISCTRCCDKYDDFEMPDKVHIH
Summary
Function
Spike protein S1: attaches the virion to the cell membrane by interacting with host receptor, initiating the infection.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Spike protein S2': Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
Spike protein S2: mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Subunit
Homotrimer; each monomer consists of a S1 and a S2 subunit. The resulting peplomers protrude from the virus surface as spikes.
Similarity
Belongs to the betacoronaviruses spike protein family.
Feature
chain Spike protein S2
Uniprot
U5LMM7
U5LNM4
A0A4D6G1A4
A0A1W6ASU7
A0A2R2YRG8
U5NJG5
+ More
A0A2I6PIX8 A0A2R4KP93 A0A2I6PIW5 A0A5H2WTJ3 A0A482DX17 A0A482DX85 A0A4P8L6Q6 A0A384YDC0 A0A0U2NE92 A0A2D0YM48 A3EXE8 A3EXF7 S4X278 A0A0A0Q4U5 A0A0U2IY58 A0A1C8EAZ2 A0A2P1IT40 A0A023VZD9 A0A2P1IT75 A0A2P1IT43 A0A493R163 A0A2H4TEF0 A0A291I635 A0A140AZ05 A0A2D0YAP4 A0A0U2NY03 A0A168DLI3 A0A2P1IT46 K0BRG7 A0A291I6E5 A0A411PBH3 A0A0U2NE84 A3EXD9 M4SVE7 A0A0H4JBS8 A0A0B5KTJ9 A0A0U2NXW5 A0A0A0Q4U2 A0A2D0YXA8 A0A2D0Y3M7 A0A2D0YUG7 A0A2H1YPP7 A0A384YDR2 A0A1L2DZI5 A0A2D0YCT4 A0A126G7T9 A0A0U2MMC5 A0A5C0PYB9 A0A0U2HTD9 A0A2D0YDA3 T2BBK5 A0A0A0Q7G4 A0A0U2NTX5 A0A384YG86 A0A140AZ15 T2B9T0 A0A140AYZ5 A0A141BR51 A0A2H4PWP1 A0A2D0YKN0 A0A0U2MS74 W6A8L9 A0A3T0I8D8 A0A0U2NQQ0 A0A2D0YT69 A0A1L2E0V8 A0A0U2J5U2 A0A0U2MMB6 A0A0U4AGH8 A0A0P0HJE7 A0A2D0YID8 A0A2D0YGK8 A0A0G2UN32 A0A0P0HBE6 W6A090 A0A1W5LIC4 A0A0A7E695 A0A1W5LGJ4 A0A2D0YGA8 A0A1W5LGD2 A0A0A0Q7F8 W6A0A7 A0A0U3SQE0 A0A2D0YCE0 A0A140AQ46 A0A0D3MU58 R9UCW7 A0A0U2MGJ8 A0A0U2P188 A0A291I6B7 A0A385AJU1 A0A291I6J0 A0A411PBG2 A0A1W5LGE4
A0A2I6PIX8 A0A2R4KP93 A0A2I6PIW5 A0A5H2WTJ3 A0A482DX17 A0A482DX85 A0A4P8L6Q6 A0A384YDC0 A0A0U2NE92 A0A2D0YM48 A3EXE8 A3EXF7 S4X278 A0A0A0Q4U5 A0A0U2IY58 A0A1C8EAZ2 A0A2P1IT40 A0A023VZD9 A0A2P1IT75 A0A2P1IT43 A0A493R163 A0A2H4TEF0 A0A291I635 A0A140AZ05 A0A2D0YAP4 A0A0U2NY03 A0A168DLI3 A0A2P1IT46 K0BRG7 A0A291I6E5 A0A411PBH3 A0A0U2NE84 A3EXD9 M4SVE7 A0A0H4JBS8 A0A0B5KTJ9 A0A0U2NXW5 A0A0A0Q4U2 A0A2D0YXA8 A0A2D0Y3M7 A0A2D0YUG7 A0A2H1YPP7 A0A384YDR2 A0A1L2DZI5 A0A2D0YCT4 A0A126G7T9 A0A0U2MMC5 A0A5C0PYB9 A0A0U2HTD9 A0A2D0YDA3 T2BBK5 A0A0A0Q7G4 A0A0U2NTX5 A0A384YG86 A0A140AZ15 T2B9T0 A0A140AYZ5 A0A141BR51 A0A2H4PWP1 A0A2D0YKN0 A0A0U2MS74 W6A8L9 A0A3T0I8D8 A0A0U2NQQ0 A0A2D0YT69 A0A1L2E0V8 A0A0U2J5U2 A0A0U2MMB6 A0A0U4AGH8 A0A0P0HJE7 A0A2D0YID8 A0A2D0YGK8 A0A0G2UN32 A0A0P0HBE6 W6A090 A0A1W5LIC4 A0A0A7E695 A0A1W5LGJ4 A0A2D0YGA8 A0A1W5LGD2 A0A0A0Q7F8 W6A0A7 A0A0U3SQE0 A0A2D0YCE0 A0A140AQ46 A0A0D3MU58 R9UCW7 A0A0U2MGJ8 A0A0U2P188 A0A291I6B7 A0A385AJU1 A0A291I6J0 A0A411PBG2 A0A1W5LGE4
Pubmed
EMBL
KC545386
AGX27810.1
KC545383
AGX27799.1
MK679660
QCC20713.1
+ More
KX574227 ARJ34226.1 MF593268 ATQ39390.1 KC869678 AGY29650.2 MG596803 AUM60024.1 MG021452 AVV62537.1 MG596802 AUM60014.1 LC469308 BBJ36008.1 MK564475 QBM11748.1 MK564474 QBM11737.1 MK858160 MK858161 MK858162 QCQ28832.1 QCQ28833.1 QCQ28834.1 MG912604 AVK87417.1 KT806010 ALJ54490.1 MF598638 ASU90406.1 EF065511 ABN10893.1 EF065512 ABN10902.1 KC522096 AGP04935.1 KM027284 MH013216 AID55095.1 AVV61900.1 KT805975 ALJ54455.1 KT877350 ALL26396.1 MG923468 AVN89313.1 KJ713299 AHY22565.1 MG923470 MG923471 AVN89334.1 AVN89344.1 MG923466 MG923467 AVN89291.1 AVN89302.1 MH978886 MH978887 QBF44113.1 QBF44114.1 MG366881 MG366882 MG366883 MG912595 MG912596 MG912598 MG912599 MG912600 MG912601 MG912602 MG912603 MG912605 MG366483 MH310910 MH310911 ATY74392.1 ATY74403.1 ATY74414.1 AVK87318.1 AVK87329.1 AVK87351.1 AVK87362.1 AVK87373.1 AVK87384.1 AVK87395.1 AVK87406.1 AVK87428.1 AXN73359.1 AXN92238.1 AXN92249.1 MG011347 MK858164 ATG84756.1 QCQ28836.1 KT868875 KX034094 KX034095 ALK80291.1 ANC28645.1 ANC28656.1 MF598679 ASU90857.1 KT806020 ALJ54500.1 KX034097 ANC28678.1 MG923469 AVN89324.1 JX869059 AFS88936.1 MG011357 ATG84866.1 MK462249 QBF80521.1 KT805994 ALJ54474.1 EF065510 ABN10884.1 KC776174 AGH58717.1 KP405225 KP405227 KT805988 MF598616 MG011341 MH822886 MK462250 AKO69634.1 AKO69636.1 ALJ54468.1 ASU90164.1 ATG84690.1 AYM48030.1 QBF80532.1 KP719931 KP719932 AJG44102.1 AJG44113.1 KT805972 KT806055 ALJ54452.1 ALW82753.1 KM027279 KT368824 AID55090.1 ALA49341.1 MF598648 MF598667 MF598696 ASU90516.1 ASU90725.1 ASU91043.1 KY673148 KY688114 MF598636 MF598639 MF598647 MF598681 AQZ41285.1 AQZ41306.1 ASU90384.1 ASU90417.1 ASU90505.1 ASU90879.1 MF598635 MF598637 ASU90373.1 ASU90395.1 MF598597 ASU89955.1 MG912597 AVK87340.1 KX108943 ANI69889.1 MF598641 ASU90439.1 KT374050 KT374051 ALB08246.1 ALB08257.1 KT805992 KY688119 ALJ54472.1 AQZ41322.1 MN365232 MN365233 QEJ82215.1 QEJ82226.1 KT805968 KT805985 KT805986 ALJ54448.1 ALJ54465.1 ALJ54466.1 MF598651 ASU90549.1 KF600634 AGV08492.1 KM027288 AID55099.1 KT806028 ALJ54508.1 MG912606 AVK87439.1 KT868872 KT868876 ALK80261.1 ALK80301.1 KF600652 KT806040 AGV08584.1 ALJ54520.1 KT182954 KT182955 KT326819 KT374052 KT374053 KT374054 KT374055 KT868865 KT868866 KT868867 KT868868 KT868869 KT868873 KT868874 KU308549 KX034096 KX034098 KX034099 KX034100 AKN11072.1 AKN11073.1 AKS48062.1 ALB08267.1 ALB08278.1 ALB08289.1 ALB08300.1 ALK80192.1 ALK80202.1 ALK80212.1 ALK80222.1 ALK80232.1 ALK80271.1 ALK80281.1 AML60270.1 ANC28667.1 ANC28689.1 ANC28700.1 ANC28711.1 KT182956 KT182957 AKN11074.1 AKN11075.1 MF679173 ATW75479.1 MF598674 ASU90802.1 KT805971 ALJ54451.1 KJ156881 AHI48550.1 MK280984 AZU90731.1 KT805966 ALJ54446.1 MF598693 ASU91010.1 KX108944 ANI69900.1 KT806038 ALJ54518.1 KT805976 ALJ54456.1 KT806049 ALW82691.1 KR912196 ALJ76286.1 MF598614 ASU90142.1 MF598608 MF598650 MF598700 ASU90076.1 ASU90538.1 ASU91087.1 KR011263 KR011264 KR011265 KR011266 KT805961 KT805962 KT805963 AKI29255.1 AKI29265.1 AKI29275.1 AKI29284.1 ALJ54441.1 ALJ54442.1 ALJ54443.1 KT368828 KT368843 KT368844 KT368845 KT368849 KT368850 KT368851 KT368855 KT368887 KT368888 KT805995 KT805997 KT805998 KT805999 KT806000 KT806013 KR912189 KR912190 KR912191 KR912192 KR912193 KR912194 KT877351 ALA49385.1 ALA49550.1 ALA49561.1 ALA49572.1 ALA49616.1 ALA49627.1 ALA49638.1 ALA49682.1 ALA50034.1 ALA50045.1 ALJ54475.1 ALJ54477.1 ALJ54478.1 ALJ54479.1 ALJ54480.1 ALJ54493.1 ALJ76282.1 ALL26409.1 KJ156876 KU242423 KU242424 MF598663 AHI48672.1 ALT66870.1 ALT66880.1 ASU90681.1 KX154689 MG757604 ANF29217.1 AXN73514.1 KP209306 KP209308 KP209309 KP209310 KP209311 KP209313 KY673147 KY581686 KY581688 KY581689 KY581690 KY581691 KY581692 KY581696 KY581697 KY581700 AIY60518.1 AIY60538.1 AIY60548.1 AIY60558.1 AIY60568.1 AIY60588.1 AQZ41283.1 ASU45719.1 ASU45741.1 ASU45752.1 ASU45763.1 ASU45774.1 ASU45785.1 ASU45829.1 ASU45840.1 ASU45873.1 KX154690 MG011345 MG011351 ANF29228.1 ATG84734.1 ATG84800.1 MF598629 MF598640 MF598658 MF598689 ASU90307.1 ASU90428.1 ASU90626.1 ASU90966.1 KX154686 KX154692 ANF29184.1 ANF29250.1 KM027273 AID55084.1 KJ156905 AHI48692.1 KT806054 ALW82742.1 MF598631 ASU90329.1 KT374056 ALB08311.1 KJ361503 AIZ74450.1 KF192507 AGN52936.1 KT806016 ALJ54496.1 KT806021 ALJ54501.1 MG011354 ATG84833.1 MH310909 AXN92228.1 MG011358 ATG84877.1 MK462251 QBF80543.1 KX154687 ANF29195.1
KX574227 ARJ34226.1 MF593268 ATQ39390.1 KC869678 AGY29650.2 MG596803 AUM60024.1 MG021452 AVV62537.1 MG596802 AUM60014.1 LC469308 BBJ36008.1 MK564475 QBM11748.1 MK564474 QBM11737.1 MK858160 MK858161 MK858162 QCQ28832.1 QCQ28833.1 QCQ28834.1 MG912604 AVK87417.1 KT806010 ALJ54490.1 MF598638 ASU90406.1 EF065511 ABN10893.1 EF065512 ABN10902.1 KC522096 AGP04935.1 KM027284 MH013216 AID55095.1 AVV61900.1 KT805975 ALJ54455.1 KT877350 ALL26396.1 MG923468 AVN89313.1 KJ713299 AHY22565.1 MG923470 MG923471 AVN89334.1 AVN89344.1 MG923466 MG923467 AVN89291.1 AVN89302.1 MH978886 MH978887 QBF44113.1 QBF44114.1 MG366881 MG366882 MG366883 MG912595 MG912596 MG912598 MG912599 MG912600 MG912601 MG912602 MG912603 MG912605 MG366483 MH310910 MH310911 ATY74392.1 ATY74403.1 ATY74414.1 AVK87318.1 AVK87329.1 AVK87351.1 AVK87362.1 AVK87373.1 AVK87384.1 AVK87395.1 AVK87406.1 AVK87428.1 AXN73359.1 AXN92238.1 AXN92249.1 MG011347 MK858164 ATG84756.1 QCQ28836.1 KT868875 KX034094 KX034095 ALK80291.1 ANC28645.1 ANC28656.1 MF598679 ASU90857.1 KT806020 ALJ54500.1 KX034097 ANC28678.1 MG923469 AVN89324.1 JX869059 AFS88936.1 MG011357 ATG84866.1 MK462249 QBF80521.1 KT805994 ALJ54474.1 EF065510 ABN10884.1 KC776174 AGH58717.1 KP405225 KP405227 KT805988 MF598616 MG011341 MH822886 MK462250 AKO69634.1 AKO69636.1 ALJ54468.1 ASU90164.1 ATG84690.1 AYM48030.1 QBF80532.1 KP719931 KP719932 AJG44102.1 AJG44113.1 KT805972 KT806055 ALJ54452.1 ALW82753.1 KM027279 KT368824 AID55090.1 ALA49341.1 MF598648 MF598667 MF598696 ASU90516.1 ASU90725.1 ASU91043.1 KY673148 KY688114 MF598636 MF598639 MF598647 MF598681 AQZ41285.1 AQZ41306.1 ASU90384.1 ASU90417.1 ASU90505.1 ASU90879.1 MF598635 MF598637 ASU90373.1 ASU90395.1 MF598597 ASU89955.1 MG912597 AVK87340.1 KX108943 ANI69889.1 MF598641 ASU90439.1 KT374050 KT374051 ALB08246.1 ALB08257.1 KT805992 KY688119 ALJ54472.1 AQZ41322.1 MN365232 MN365233 QEJ82215.1 QEJ82226.1 KT805968 KT805985 KT805986 ALJ54448.1 ALJ54465.1 ALJ54466.1 MF598651 ASU90549.1 KF600634 AGV08492.1 KM027288 AID55099.1 KT806028 ALJ54508.1 MG912606 AVK87439.1 KT868872 KT868876 ALK80261.1 ALK80301.1 KF600652 KT806040 AGV08584.1 ALJ54520.1 KT182954 KT182955 KT326819 KT374052 KT374053 KT374054 KT374055 KT868865 KT868866 KT868867 KT868868 KT868869 KT868873 KT868874 KU308549 KX034096 KX034098 KX034099 KX034100 AKN11072.1 AKN11073.1 AKS48062.1 ALB08267.1 ALB08278.1 ALB08289.1 ALB08300.1 ALK80192.1 ALK80202.1 ALK80212.1 ALK80222.1 ALK80232.1 ALK80271.1 ALK80281.1 AML60270.1 ANC28667.1 ANC28689.1 ANC28700.1 ANC28711.1 KT182956 KT182957 AKN11074.1 AKN11075.1 MF679173 ATW75479.1 MF598674 ASU90802.1 KT805971 ALJ54451.1 KJ156881 AHI48550.1 MK280984 AZU90731.1 KT805966 ALJ54446.1 MF598693 ASU91010.1 KX108944 ANI69900.1 KT806038 ALJ54518.1 KT805976 ALJ54456.1 KT806049 ALW82691.1 KR912196 ALJ76286.1 MF598614 ASU90142.1 MF598608 MF598650 MF598700 ASU90076.1 ASU90538.1 ASU91087.1 KR011263 KR011264 KR011265 KR011266 KT805961 KT805962 KT805963 AKI29255.1 AKI29265.1 AKI29275.1 AKI29284.1 ALJ54441.1 ALJ54442.1 ALJ54443.1 KT368828 KT368843 KT368844 KT368845 KT368849 KT368850 KT368851 KT368855 KT368887 KT368888 KT805995 KT805997 KT805998 KT805999 KT806000 KT806013 KR912189 KR912190 KR912191 KR912192 KR912193 KR912194 KT877351 ALA49385.1 ALA49550.1 ALA49561.1 ALA49572.1 ALA49616.1 ALA49627.1 ALA49638.1 ALA49682.1 ALA50034.1 ALA50045.1 ALJ54475.1 ALJ54477.1 ALJ54478.1 ALJ54479.1 ALJ54480.1 ALJ54493.1 ALJ76282.1 ALL26409.1 KJ156876 KU242423 KU242424 MF598663 AHI48672.1 ALT66870.1 ALT66880.1 ASU90681.1 KX154689 MG757604 ANF29217.1 AXN73514.1 KP209306 KP209308 KP209309 KP209310 KP209311 KP209313 KY673147 KY581686 KY581688 KY581689 KY581690 KY581691 KY581692 KY581696 KY581697 KY581700 AIY60518.1 AIY60538.1 AIY60548.1 AIY60558.1 AIY60568.1 AIY60588.1 AQZ41283.1 ASU45719.1 ASU45741.1 ASU45752.1 ASU45763.1 ASU45774.1 ASU45785.1 ASU45829.1 ASU45840.1 ASU45873.1 KX154690 MG011345 MG011351 ANF29228.1 ATG84734.1 ATG84800.1 MF598629 MF598640 MF598658 MF598689 ASU90307.1 ASU90428.1 ASU90626.1 ASU90966.1 KX154686 KX154692 ANF29184.1 ANF29250.1 KM027273 AID55084.1 KJ156905 AHI48692.1 KT806054 ALW82742.1 MF598631 ASU90329.1 KT374056 ALB08311.1 KJ361503 AIZ74450.1 KF192507 AGN52936.1 KT806016 ALJ54496.1 KT806021 ALJ54501.1 MG011354 ATG84833.1 MH310909 AXN92228.1 MG011358 ATG84877.1 MK462251 QBF80543.1 KX154687 ANF29195.1
Proteomes
UP000095937
UP000101546
UP000202992
UP000145931
UP000143769
UP000100668
+ More
UP000105499 UP000145271 UP000167218 UP000129370 UP000112279 UP000164987 UP000108469 UP000131416 UP000174176 UP000117213 UP000138672 UP000103178 UP000113724 UP000096281 UP000148810 UP000097149 UP000098933 UP000105311 UP000106595 UP000109122 UP000126763 UP000128076 UP000140611 UP000147342 UP000105272 UP000146591 UP000102098 UP000110604 UP000120083 UP000165587 UP000098431 UP000104003 UP000112676 UP000126955 UP000128936 UP000136645 UP000137287 UP000146604 UP000151733 UP000159793 UP000120625 UP000174530 UP000110817 UP000118636 UP000130850 UP000142709 UP000144516 UP000159028 UP000171389 UP000149108 UP000096629 UP000104220
UP000105499 UP000145271 UP000167218 UP000129370 UP000112279 UP000164987 UP000108469 UP000131416 UP000174176 UP000117213 UP000138672 UP000103178 UP000113724 UP000096281 UP000148810 UP000097149 UP000098933 UP000105311 UP000106595 UP000109122 UP000126763 UP000128076 UP000140611 UP000147342 UP000105272 UP000146591 UP000102098 UP000110604 UP000120083 UP000165587 UP000098431 UP000104003 UP000112676 UP000126955 UP000128936 UP000136645 UP000137287 UP000146604 UP000151733 UP000159793 UP000120625 UP000174530 UP000110817 UP000118636 UP000130850 UP000142709 UP000144516 UP000159028 UP000171389 UP000149108 UP000096629 UP000104220
Interpro
SUPFAM
SSF143587
SSF143587
Gene 3D
ProteinModelPortal
U5LMM7
U5LNM4
A0A4D6G1A4
A0A1W6ASU7
A0A2R2YRG8
U5NJG5
+ More
A0A2I6PIX8 A0A2R4KP93 A0A2I6PIW5 A0A5H2WTJ3 A0A482DX17 A0A482DX85 A0A4P8L6Q6 A0A384YDC0 A0A0U2NE92 A0A2D0YM48 A3EXE8 A3EXF7 S4X278 A0A0A0Q4U5 A0A0U2IY58 A0A1C8EAZ2 A0A2P1IT40 A0A023VZD9 A0A2P1IT75 A0A2P1IT43 A0A493R163 A0A2H4TEF0 A0A291I635 A0A140AZ05 A0A2D0YAP4 A0A0U2NY03 A0A168DLI3 A0A2P1IT46 K0BRG7 A0A291I6E5 A0A411PBH3 A0A0U2NE84 A3EXD9 M4SVE7 A0A0H4JBS8 A0A0B5KTJ9 A0A0U2NXW5 A0A0A0Q4U2 A0A2D0YXA8 A0A2D0Y3M7 A0A2D0YUG7 A0A2H1YPP7 A0A384YDR2 A0A1L2DZI5 A0A2D0YCT4 A0A126G7T9 A0A0U2MMC5 A0A5C0PYB9 A0A0U2HTD9 A0A2D0YDA3 T2BBK5 A0A0A0Q7G4 A0A0U2NTX5 A0A384YG86 A0A140AZ15 T2B9T0 A0A140AYZ5 A0A141BR51 A0A2H4PWP1 A0A2D0YKN0 A0A0U2MS74 W6A8L9 A0A3T0I8D8 A0A0U2NQQ0 A0A2D0YT69 A0A1L2E0V8 A0A0U2J5U2 A0A0U2MMB6 A0A0U4AGH8 A0A0P0HJE7 A0A2D0YID8 A0A2D0YGK8 A0A0G2UN32 A0A0P0HBE6 W6A090 A0A1W5LIC4 A0A0A7E695 A0A1W5LGJ4 A0A2D0YGA8 A0A1W5LGD2 A0A0A0Q7F8 W6A0A7 A0A0U3SQE0 A0A2D0YCE0 A0A140AQ46 A0A0D3MU58 R9UCW7 A0A0U2MGJ8 A0A0U2P188 A0A291I6B7 A0A385AJU1 A0A291I6J0 A0A411PBG2 A0A1W5LGE4
A0A2I6PIX8 A0A2R4KP93 A0A2I6PIW5 A0A5H2WTJ3 A0A482DX17 A0A482DX85 A0A4P8L6Q6 A0A384YDC0 A0A0U2NE92 A0A2D0YM48 A3EXE8 A3EXF7 S4X278 A0A0A0Q4U5 A0A0U2IY58 A0A1C8EAZ2 A0A2P1IT40 A0A023VZD9 A0A2P1IT75 A0A2P1IT43 A0A493R163 A0A2H4TEF0 A0A291I635 A0A140AZ05 A0A2D0YAP4 A0A0U2NY03 A0A168DLI3 A0A2P1IT46 K0BRG7 A0A291I6E5 A0A411PBH3 A0A0U2NE84 A3EXD9 M4SVE7 A0A0H4JBS8 A0A0B5KTJ9 A0A0U2NXW5 A0A0A0Q4U2 A0A2D0YXA8 A0A2D0Y3M7 A0A2D0YUG7 A0A2H1YPP7 A0A384YDR2 A0A1L2DZI5 A0A2D0YCT4 A0A126G7T9 A0A0U2MMC5 A0A5C0PYB9 A0A0U2HTD9 A0A2D0YDA3 T2BBK5 A0A0A0Q7G4 A0A0U2NTX5 A0A384YG86 A0A140AZ15 T2B9T0 A0A140AYZ5 A0A141BR51 A0A2H4PWP1 A0A2D0YKN0 A0A0U2MS74 W6A8L9 A0A3T0I8D8 A0A0U2NQQ0 A0A2D0YT69 A0A1L2E0V8 A0A0U2J5U2 A0A0U2MMB6 A0A0U4AGH8 A0A0P0HJE7 A0A2D0YID8 A0A2D0YGK8 A0A0G2UN32 A0A0P0HBE6 W6A090 A0A1W5LIC4 A0A0A7E695 A0A1W5LGJ4 A0A2D0YGA8 A0A1W5LGD2 A0A0A0Q7F8 W6A0A7 A0A0U3SQE0 A0A2D0YCE0 A0A140AQ46 A0A0D3MU58 R9UCW7 A0A0U2MGJ8 A0A0U2P188 A0A291I6B7 A0A385AJU1 A0A291I6J0 A0A411PBG2 A0A1W5LGE4
PDB
5X5F
E-value=0,
Score=3563
Ontologies
GO
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0061025 P:membrane fusion
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0020002 C:host cell plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0016021 C:integral component of membrane
GO:0061025 P:membrane fusion
GO:0046813 P:receptor-mediated virion attachment to host cell
GO:0019064 P:fusion of virus membrane with host plasma membrane
GO:0020002 C:host cell plasma membrane
GO:0075509 P:endocytosis involved in viral entry into host cell
GO:0039654 P:fusion of virus membrane with host endosome membrane
GO:0009405 P:pathogenesis
GO:0055036 C:virion membrane
GO:0044173 C:host cell endoplasmic reticulum-Golgi intermediate compartment membrane
Subcellular Location
From MSLVP
Capsid
From Uniprot
Virion membrane
Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
Host cell membrane Accumulates in the endoplasmic reticulum-Golgi intermediate compartment, where it participates in virus particle assembly. Some S oligomers are transported to the host plasma membrane, where they may mediate cell-cell fusion. With evidence from 1 publications.
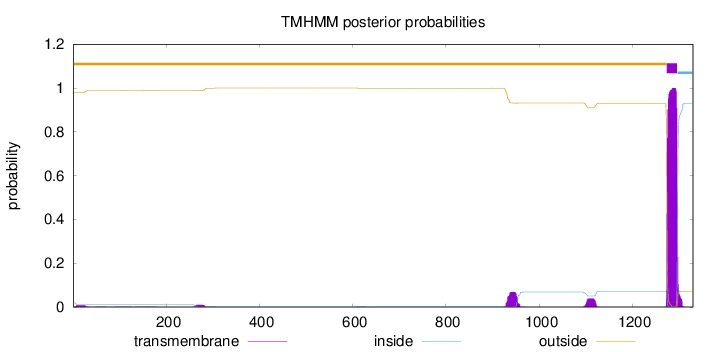
Topology
Length:
1330
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.98302
Exp number, first 60 AAs:
0.18167
Total prob of N-in:
0.01923
outside
1 - 1273
TMhelix
1274 - 1296
inside
1297 - 1330
Population Genetic Test Statistics
Pi
0.3929321
Theta
0.4414017
Tajima's D
-0.6712777
CLR
14.80375
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 | |||
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 | |||
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 | |||
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 | |||
| CHINA_MURINE_2012_NC_026011 | |||
| GERMANY_ERINACEINAE_2012_NC_022643 |
Protein |
YP_008719932.1 |
|
| GERMANY_ERINACEINAE_2012_NC_039207 |
Protein |
YP_009513010.1 |
|
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.