Strain
EUROPE_MURINE_2004_AY700211
(Region: Europe; Strain: Murine hepatitis virus strain A59, complete genome.; Date: 2004)
Gene
membrane protein
Description
Annotated in NCBI,
membrane protein
Location
GenBank Accession
Full name
Membrane protein
Alternative Name
E1 glycoprotein
Matrix glycoprotein
Membrane glycoprotein
Matrix glycoprotein
Membrane glycoprotein
Sequence
CDS
ATGAGTAGTACTACTCAGGCCCCAGAGCCCGTCTATCAATGGACCGCCGACGAGGCAGTTCAATTCCTTAAGGAATGGAACTTCTCGTTGGGCATTATACTACTCTTTATTACTATCATACTACAGTTCGGTTACACGAGCCGTAGCATGTTTATTTATGTTGTGAAAATGATAATCTTGTGGTTAATGTGGCCACTGACTATTGTTTTGTGTATTTTCAATTGCGTGTATGCGCTAAATAATGTGTATCTTGGATTTTCTATAGTGTTTACTATAGTGTCCATTGTAATCTGGATTATGTATTTTGTTAATAGCATAAGGTTGTTTATCAGGACTGGTAGCTGGTGGAGCTTCAACCCCGAAACAAACAACCTTATGTGTATAGATATGAAAGGTACCGTGTATGTTAGACCCATTATTGAGGATTACCATACACTAACAGCCACTATTATTCGTGGCCACCTCTACATGCAAGGTGTTAAGCTAGGCACCGGTTTCTCTTTGTCTGACTTGCCCGCTTATGTTACAGTTGCTAAGGTGTCACACCTTTGCACTTATAAGCGCGCATTCTTAGACAAGGTAGACGGTGTTAGCGGTTTTGCTGTTTATGTGAAGTCCAAGGTCGGAAATTACCGACTGCCCTCAAACAAACCGAGTGGCGCGGACACCGCATTGTTGAGAACCTAA
Protein
MSSTTQAPEPVYQWTADEAVQFLKEWNFSLGIILLFITIILQFGYTSRSMFIYVVKMIILWLMWPLTIVLCIFNCVYALNNVYLGFSIVFTIVSIVIWIMYFVNSIRLFIRTGSWWSFNPETNNLMCIDMKGTVYVRPIIEDYHTLTATIIRGHLYMQGVKLGTGFSLSDLPAYVTVAKVSHLCTYKRAFLDKVDGVSGFAVYVKSKVGNYRLPSNKPSGADTALLRT
Summary
Function
Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins.
Subunit
Homomultimer. Interacts with envelope E protein in the budding compartment of the host cell, which is located between endoplasmic reticulum and the Golgi complex. Forms a complex with HE and S proteins. Interacts with nucleocapsid N protein. This interaction probably participates in RNA packaging into the virus.
Homomultimer. Interacts with envelope E protein in the budding compartment of the host cell, which is located between endoplasmic reticulum and the Golgi complex. Forms a complex with HE and S proteins. Interacts with nucleocapsid N protein. This interaction probably participates in RNA packaging into the virus (By similarity).
Homomultimer. Interacts with envelope E protein in the budding compartment of the host cell, which is located between endoplasmic reticulum and the Golgi complex. Forms a complex with HE and S proteins. Interacts with nucleocapsid N protein. This interaction probably participates in RNA packaging into the virus (By similarity).
Similarity
Belongs to the betacoronaviruses M protein family.
Belongs to the coronaviruses M protein family.
Belongs to the alphacoronaviruses M protein family.
Belongs to the coronaviruses M protein family.
Belongs to the alphacoronaviruses M protein family.
Keywords
Glycoprotein
Host Golgi apparatus
Host membrane
Host-virus interaction
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Viral envelope protein
Viral immunoevasion
Viral matrix protein
Virion
Feature
chain Membrane protein
Uniprot
C0KYT4
C0KYR4
C0KYY2
C0KZ03
Q9J3E3
S5YGC7
+ More
A7U548 C0KYW5 Q77NM8 A0A2D1CEC5 C3VPF2 P03415 A0A2S0SZ14 C0KYX3 A0A2D1CEF4 H9BZY2 D7PJY4 Q5ICW8 C0KYZ3 C0KYS5 C0KYU4 C0KYV5 A0A513RAU2 Q9J3F0 D4QFK5 Q9JEB4 Q7TG51 I7AB50 P08549 Q9QD00 I7AB57 A0A513RAU4 C6GHS5 I1TMG6 Q9IKC7 V5N7E5 Q9WCD2 I1TMH6 Q9WCD1 A0A2H4MXA4 A0A096XNI0 A0A2H4MWZ9 A0A2H4N019 A0A096XNG2 A0A096XNH8 A0A2H4MXY2 A0A3G3NH91 A0A3G3NH08 A0A2P1IQA5 A0A2H4MZ01 A0A3G3NH59 C6GHN0 A4ZU79 C6GHP2 A4ZU13 A4ZU68 C6GHQ3 A4ZTY0 B7U2P8 A4ZU46 A4ZTZ1 A4ZU35 Q06BD5 A4ZU57 A4ZU02 Q4U3R5 Q8V433 B7U2S0 B7U2N8 B7U2M7 C6GHL8 B7U2L6 A3FEW1 Q0PL34 A3FEW0 Q4U3R0 A0A191URC8 B7U2Q9 P69600 P69599 A0A0A7V404 Q91A23 A0A3S7GYH1 Q6W359 Q4U3R7 A4ZU24 H9AA58 A0A096XNF0 A0A0C5B155 B8RIR2 D0EY98 A0A3G3NGZ2 H9AA37 X2JFA7 A0A0U2GR22 P69705 P69704 P69703 C6GHR5 A7BKC7
A7U548 C0KYW5 Q77NM8 A0A2D1CEC5 C3VPF2 P03415 A0A2S0SZ14 C0KYX3 A0A2D1CEF4 H9BZY2 D7PJY4 Q5ICW8 C0KYZ3 C0KYS5 C0KYU4 C0KYV5 A0A513RAU2 Q9J3F0 D4QFK5 Q9JEB4 Q7TG51 I7AB50 P08549 Q9QD00 I7AB57 A0A513RAU4 C6GHS5 I1TMG6 Q9IKC7 V5N7E5 Q9WCD2 I1TMH6 Q9WCD1 A0A2H4MXA4 A0A096XNI0 A0A2H4MWZ9 A0A2H4N019 A0A096XNG2 A0A096XNH8 A0A2H4MXY2 A0A3G3NH91 A0A3G3NH08 A0A2P1IQA5 A0A2H4MZ01 A0A3G3NH59 C6GHN0 A4ZU79 C6GHP2 A4ZU13 A4ZU68 C6GHQ3 A4ZTY0 B7U2P8 A4ZU46 A4ZTZ1 A4ZU35 Q06BD5 A4ZU57 A4ZU02 Q4U3R5 Q8V433 B7U2S0 B7U2N8 B7U2M7 C6GHL8 B7U2L6 A3FEW1 Q0PL34 A3FEW0 Q4U3R0 A0A191URC8 B7U2Q9 P69600 P69599 A0A0A7V404 Q91A23 A0A3S7GYH1 Q6W359 Q4U3R7 A4ZU24 H9AA58 A0A096XNF0 A0A0C5B155 B8RIR2 D0EY98 A0A3G3NGZ2 H9AA37 X2JFA7 A0A0U2GR22 P69705 P69704 P69703 C6GHR5 A7BKC7
Pubmed
10438865
15709029
17079303
11582515
16341254
20631137
+ More
17041219 8392595 6325918 6331107 9426441 1400501 10438834 10799570 1629209 20519394 15507445 12970410 12663803 10725548 11502093 12784855 3748812 10544100 10933723 10882653 10551451 30285857 25463600 29508550 17344285 17434558 18842722 17459444 16604443 11714968 27274850 9778786 25552712 22398294 19162098 24655427 26678874 11774548 3029965 3434434 7966615 9371586 1706882 17374765
17041219 8392595 6325918 6331107 9426441 1400501 10438834 10799570 1629209 20519394 15507445 12970410 12663803 10725548 11502093 12784855 3748812 10544100 10933723 10882653 10551451 30285857 25463600 29508550 17344285 17434558 18842722 17459444 16604443 11714968 27274850 9778786 25552712 22398294 19162098 24655427 26678874 11774548 3029965 3434434 7966615 9371586 1706882 17374765
EMBL
FJ647220
ACN89707.1
FJ647218
ACN89692.1
FJ647225
ACN89755.1
+ More
FJ647227 ACN89781.1 AF208067 AY700211 AAF69348.1 AAU06360.1 KF268336 KF268337 KF268338 KF268339 AGT17720.1 AGT17731.1 AGT17742.1 AGT17753.1 EF682498 ABS87268.1 FJ647223 ACN89740.1 AF207902 AAF68927.1 MF618252 ATN37891.1 FJ884686 FJ884687 ACO72887.1 X00509 M25894 AF029248 MF416379 AWB14617.1 FJ647224 ACN89749.1 MF618253 ATN37901.1 JQ173883 AFD97611.1 GU593319 ADI59792.1 AY771998 AAW47244.1 FJ647226 ACN89768.1 FJ647219 ACN89700.1 FJ647221 ACN89721.1 FJ647222 ACN89725.1 MH687884 MH687885 QCI55925.1 QCI55926.1 AF208066 AAF69337.1 AB551247 BAJ04700.1 AF126508 AF201929 AY102934 AAM55466.1 JX169866 AFO11511.1 X04223 AF190407 AAF05705.1 JX169867 AFO11521.1 MH687886 MH687887 MH687888 QCI55927.1 QCI55928.1 QCI55929.1 FJ938068 ACT11045.1 JF792616 AFG25754.1 AF207551 KF850449 AHA83611.1 AF088986 AAD33106.1 JF792617 AFG25764.1 AF088985 KY370043 ATP66729.1 KF294372 AID16663.1 KY370048 ATP66758.1 KY370051 ATP66775.1 KF294370 AID16651.1 KF294371 AID16657.1 KY370049 ATP66764.1 MH687970 AYR18620.1 MH687968 AYR18602.1 MG518518 AVN88327.1 KY370047 ATP66752.1 MH687971 MH687972 MH687973 MH687974 MH687975 MH687976 MH687977 MH687978 AYR18629.1 AYR18638.1 AYR18647.1 AYR18656.1 AYR18665.1 AYR18674.1 AYR18683.1 AYR18692.1 FJ938064 ACT11000.1 EF424624 ABP38331.1 FJ938065 ACT11012.1 EF424618 ABP38270.1 EF424623 ABP38325.1 FJ938066 ACT11023.1 EF424615 ABP38238.1 FJ425187 ACJ66984.1 EF424621 ABP38305.1 EF424616 ABP38250.1 EF424620 ABP38285.1 DQ915164 ABI94002.1 EF424622 ABP38308.1 EF424617 ABP38255.1 DQ016129 DQ016130 DQ016131 DQ016132 DQ016134 AAY33933.1 AAY33936.1 AAY33939.1 AAY33942.1 AAY33948.1 AF391542 FJ425189 ACJ67005.1 FJ425186 ACJ66966.1 FJ425185 ACJ66958.1 FJ938063 ACT10988.1 FJ425184 ACJ66952.1 EF371516 ABN49621.1 DQ811784 ABG89282.1 EF371515 ABN49620.1 AH014871 AH014873 KU886219 AAY33945.1 AAY33951.1 ANJ04979.1 KU558922 KU558923 ANJ04722.1 ANJ04733.1 FJ425188 FJ425190 ACJ66992.1 AF058944 AF058943 KM349742 KM349743 KM349744 AJA91200.1 AJA91211.1 AJA91221.1 AF391541 MG757138 MG757139 MG757140 MG757141 MG757142 AVI15005.1 AVI15016.1 AVI15027.1 AVI15038.1 AVI15049.1 AY316299 FJ415324 AAQ67201.1 ACJ35488.1 AH014866 AAY33930.1 EF424619 ABP38278.1 JN874561 AFE48820.1 KF294357 AID16633.1 KM985633 AJL33726.1 EU999954 ACL13000.1 GQ918143 ACX46865.1 MH687969 AYR18611.1 JN874559 JN874562 AFE48798.1 AFE48830.1 KF906249 KF906250 KF906251 AHN64777.1 AHN64786.1 AHN64795.1 KT368891 MF593476 ALA50083.1 ATI09452.1 AF220295 M27474 U00735 FJ938067 ACT11035.1 AB354579 BAF75636.1
FJ647227 ACN89781.1 AF208067 AY700211 AAF69348.1 AAU06360.1 KF268336 KF268337 KF268338 KF268339 AGT17720.1 AGT17731.1 AGT17742.1 AGT17753.1 EF682498 ABS87268.1 FJ647223 ACN89740.1 AF207902 AAF68927.1 MF618252 ATN37891.1 FJ884686 FJ884687 ACO72887.1 X00509 M25894 AF029248 MF416379 AWB14617.1 FJ647224 ACN89749.1 MF618253 ATN37901.1 JQ173883 AFD97611.1 GU593319 ADI59792.1 AY771998 AAW47244.1 FJ647226 ACN89768.1 FJ647219 ACN89700.1 FJ647221 ACN89721.1 FJ647222 ACN89725.1 MH687884 MH687885 QCI55925.1 QCI55926.1 AF208066 AAF69337.1 AB551247 BAJ04700.1 AF126508 AF201929 AY102934 AAM55466.1 JX169866 AFO11511.1 X04223 AF190407 AAF05705.1 JX169867 AFO11521.1 MH687886 MH687887 MH687888 QCI55927.1 QCI55928.1 QCI55929.1 FJ938068 ACT11045.1 JF792616 AFG25754.1 AF207551 KF850449 AHA83611.1 AF088986 AAD33106.1 JF792617 AFG25764.1 AF088985 KY370043 ATP66729.1 KF294372 AID16663.1 KY370048 ATP66758.1 KY370051 ATP66775.1 KF294370 AID16651.1 KF294371 AID16657.1 KY370049 ATP66764.1 MH687970 AYR18620.1 MH687968 AYR18602.1 MG518518 AVN88327.1 KY370047 ATP66752.1 MH687971 MH687972 MH687973 MH687974 MH687975 MH687976 MH687977 MH687978 AYR18629.1 AYR18638.1 AYR18647.1 AYR18656.1 AYR18665.1 AYR18674.1 AYR18683.1 AYR18692.1 FJ938064 ACT11000.1 EF424624 ABP38331.1 FJ938065 ACT11012.1 EF424618 ABP38270.1 EF424623 ABP38325.1 FJ938066 ACT11023.1 EF424615 ABP38238.1 FJ425187 ACJ66984.1 EF424621 ABP38305.1 EF424616 ABP38250.1 EF424620 ABP38285.1 DQ915164 ABI94002.1 EF424622 ABP38308.1 EF424617 ABP38255.1 DQ016129 DQ016130 DQ016131 DQ016132 DQ016134 AAY33933.1 AAY33936.1 AAY33939.1 AAY33942.1 AAY33948.1 AF391542 FJ425189 ACJ67005.1 FJ425186 ACJ66966.1 FJ425185 ACJ66958.1 FJ938063 ACT10988.1 FJ425184 ACJ66952.1 EF371516 ABN49621.1 DQ811784 ABG89282.1 EF371515 ABN49620.1 AH014871 AH014873 KU886219 AAY33945.1 AAY33951.1 ANJ04979.1 KU558922 KU558923 ANJ04722.1 ANJ04733.1 FJ425188 FJ425190 ACJ66992.1 AF058944 AF058943 KM349742 KM349743 KM349744 AJA91200.1 AJA91211.1 AJA91221.1 AF391541 MG757138 MG757139 MG757140 MG757141 MG757142 AVI15005.1 AVI15016.1 AVI15027.1 AVI15038.1 AVI15049.1 AY316299 FJ415324 AAQ67201.1 ACJ35488.1 AH014866 AAY33930.1 EF424619 ABP38278.1 JN874561 AFE48820.1 KF294357 AID16633.1 KM985633 AJL33726.1 EU999954 ACL13000.1 GQ918143 ACX46865.1 MH687969 AYR18611.1 JN874559 JN874562 AFE48798.1 AFE48830.1 KF906249 KF906250 KF906251 AHN64777.1 AHN64786.1 AHN64795.1 KT368891 MF593476 ALA50083.1 ATI09452.1 AF220295 M27474 U00735 FJ938067 ACT11035.1 AB354579 BAF75636.1
Proteomes
UP000098534
UP000151686
UP000119015
UP000138757
UP000113498
UP000155907
+ More
UP000112338 UP000164259 UP000171340 UP000171975 UP000159025 UP000163097 UP000180349 UP000180350 UP000007192 UP000167944 UP000118088 UP000117254 UP000148943 UP000158592 UP000133456 UP000142874 UP000143567 UP000154289 UP000139707 UP000114687 UP000007193 UP000149957 UP000173347 UP000175084 UP000161770 UP000136745 UP000147178 UP000139966 UP000173415 UP000163913 UP000162935 UP000121953 UP000158327 UP000136116 UP000127099 UP000170940 UP000154814 UP000139235 UP000151273 UP000008571 UP000133443 UP000140344 UP000104294 UP000172225 UP000174569 UP000147981 UP000102696 UP000148210 UP000144342 UP000153628 UP000174571 UP000008570 UP000158296 UP000134650 UP000125269 UP000107676 UP000145001 UP000121258 UP000145507 UP000149260 UP000158231 UP000008572 UP000007554 UP000164707 UP000168103
UP000112338 UP000164259 UP000171340 UP000171975 UP000159025 UP000163097 UP000180349 UP000180350 UP000007192 UP000167944 UP000118088 UP000117254 UP000148943 UP000158592 UP000133456 UP000142874 UP000143567 UP000154289 UP000139707 UP000114687 UP000007193 UP000149957 UP000173347 UP000175084 UP000161770 UP000136745 UP000147178 UP000139966 UP000173415 UP000163913 UP000162935 UP000121953 UP000158327 UP000136116 UP000127099 UP000170940 UP000154814 UP000139235 UP000151273 UP000008571 UP000133443 UP000140344 UP000104294 UP000172225 UP000174569 UP000147981 UP000102696 UP000148210 UP000144342 UP000153628 UP000174571 UP000008570 UP000158296 UP000134650 UP000125269 UP000107676 UP000145001 UP000121258 UP000145507 UP000149260 UP000158231 UP000008572 UP000007554 UP000164707 UP000168103
PRIDE
Pfam
PF01635
Corona_M
Interpro
IPR002574
Corona_M
ProteinModelPortal
C0KYT4
C0KYR4
C0KYY2
C0KZ03
Q9J3E3
S5YGC7
+ More
A7U548 C0KYW5 Q77NM8 A0A2D1CEC5 C3VPF2 P03415 A0A2S0SZ14 C0KYX3 A0A2D1CEF4 H9BZY2 D7PJY4 Q5ICW8 C0KYZ3 C0KYS5 C0KYU4 C0KYV5 A0A513RAU2 Q9J3F0 D4QFK5 Q9JEB4 Q7TG51 I7AB50 P08549 Q9QD00 I7AB57 A0A513RAU4 C6GHS5 I1TMG6 Q9IKC7 V5N7E5 Q9WCD2 I1TMH6 Q9WCD1 A0A2H4MXA4 A0A096XNI0 A0A2H4MWZ9 A0A2H4N019 A0A096XNG2 A0A096XNH8 A0A2H4MXY2 A0A3G3NH91 A0A3G3NH08 A0A2P1IQA5 A0A2H4MZ01 A0A3G3NH59 C6GHN0 A4ZU79 C6GHP2 A4ZU13 A4ZU68 C6GHQ3 A4ZTY0 B7U2P8 A4ZU46 A4ZTZ1 A4ZU35 Q06BD5 A4ZU57 A4ZU02 Q4U3R5 Q8V433 B7U2S0 B7U2N8 B7U2M7 C6GHL8 B7U2L6 A3FEW1 Q0PL34 A3FEW0 Q4U3R0 A0A191URC8 B7U2Q9 P69600 P69599 A0A0A7V404 Q91A23 A0A3S7GYH1 Q6W359 Q4U3R7 A4ZU24 H9AA58 A0A096XNF0 A0A0C5B155 B8RIR2 D0EY98 A0A3G3NGZ2 H9AA37 X2JFA7 A0A0U2GR22 P69705 P69704 P69703 C6GHR5 A7BKC7
A7U548 C0KYW5 Q77NM8 A0A2D1CEC5 C3VPF2 P03415 A0A2S0SZ14 C0KYX3 A0A2D1CEF4 H9BZY2 D7PJY4 Q5ICW8 C0KYZ3 C0KYS5 C0KYU4 C0KYV5 A0A513RAU2 Q9J3F0 D4QFK5 Q9JEB4 Q7TG51 I7AB50 P08549 Q9QD00 I7AB57 A0A513RAU4 C6GHS5 I1TMG6 Q9IKC7 V5N7E5 Q9WCD2 I1TMH6 Q9WCD1 A0A2H4MXA4 A0A096XNI0 A0A2H4MWZ9 A0A2H4N019 A0A096XNG2 A0A096XNH8 A0A2H4MXY2 A0A3G3NH91 A0A3G3NH08 A0A2P1IQA5 A0A2H4MZ01 A0A3G3NH59 C6GHN0 A4ZU79 C6GHP2 A4ZU13 A4ZU68 C6GHQ3 A4ZTY0 B7U2P8 A4ZU46 A4ZTZ1 A4ZU35 Q06BD5 A4ZU57 A4ZU02 Q4U3R5 Q8V433 B7U2S0 B7U2N8 B7U2M7 C6GHL8 B7U2L6 A3FEW1 Q0PL34 A3FEW0 Q4U3R0 A0A191URC8 B7U2Q9 P69600 P69599 A0A0A7V404 Q91A23 A0A3S7GYH1 Q6W359 Q4U3R7 A4ZU24 H9AA58 A0A096XNF0 A0A0C5B155 B8RIR2 D0EY98 A0A3G3NGZ2 H9AA37 X2JFA7 A0A0U2GR22 P69705 P69704 P69703 C6GHR5 A7BKC7
Ontologies
GO
GO:0030683 P:evasion or tolerance by virus of host immune response
GO:0039660 F:structural constituent of virion
GO:0055036 C:virion membrane
GO:0044178 C:host cell Golgi membrane
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0019058 P:viral life cycle
GO:0044177 C:host cell Golgi apparatus
GO:0039660 F:structural constituent of virion
GO:0055036 C:virion membrane
GO:0044178 C:host cell Golgi membrane
GO:0019031 C:viral envelope
GO:0016021 C:integral component of membrane
GO:0019058 P:viral life cycle
GO:0044177 C:host cell Golgi apparatus
Subcellular Location
From MSLVP
Host nucleus. Host cytoplasm.
From Uniprot
Virion membrane
Largely embedded in the lipid bilayer. With evidence from 1 publications.
Host Golgi apparatus membrane Largely embedded in the lipid bilayer. With evidence from 1 publications.
Host Golgi apparatus membrane Largely embedded in the lipid bilayer. With evidence from 1 publications.
Topology
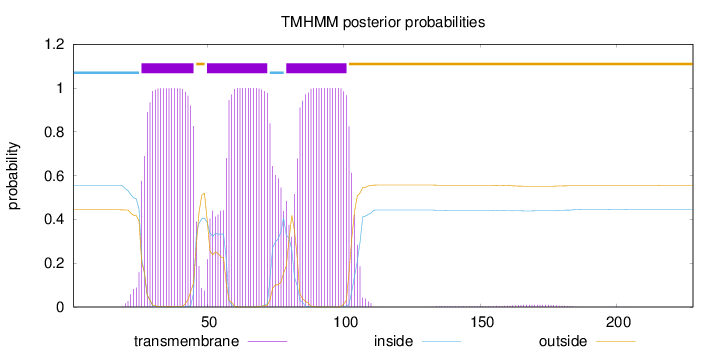
Length:
228
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
64.7462799999999
Exp number, first 60 AAs:
26.28326
Total prob of N-in:
0.55612
POSSIBLE N-term signal
sequence
inside
1 - 25
TMhelix
26 - 45
outside
46 - 49
TMhelix
50 - 72
inside
73 - 78
TMhelix
79 - 101
outside
102 - 228
Population Genetic Test Statistics
Pi
0.2132888
Theta
0.3348037
Tajima's D
-1.0476155
CLR
3.554251
Interpretation
No evidence of Selection
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 | |||
| CHINA_RABBIT_2006_NC_017083 |
Protein |
YP_005454248.1 |
|
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 |
Protein |
AAU06360.1 |
|
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 | |||
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 |
Protein |
AAT84357.1 |
|
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 |
Protein |
YP_173241.1 |
|
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 |
Protein |
NP_150082.1 |
|
| CHINA_MURINE_2012_NC_026011 |
Protein |
YP_009113029.1 |
|
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
