Strain
USA_MURINE_1997_NC_001846
(Region: USA: NY; Strain: Mouse hepatitis virus strain MHV-A59 C12 mutant, complete genome.; Date: 1997)
Gene
nucleocapsid protein
Description
Annotated in NCBI,
nucleocapsid protein
Location
GenBank Accession
Full name
Nucleoprotein
Alternative Name
Nucleocapsid protein
Sequence
CDS
ATGTCTTTTGTTCCTGGGCAAGAAAATGCCGGTGGCAGAAGCTCCTCTGTAAACCGCGCTGGTAATGGAATCCTCAAGAAGACCACTTGGGCTGACCAAACCGAGCGTGGACCAAATAATCAAAATAGAGGCAGAAGGAATCAGCCAAAGCAGACTGCAACTACTCAACCCAACTCCGGGAGTGTGGTTCCCCATTACTCCTGGTTTTCTGGCATTACCCAGTTCCAAAAGGGAAAGGAGTTTCAGTTTGCAGAAGGACAAGGAGTGCCTATTGCCAATGGAATCCCCGCTTCAGAGCAAAAGGGATATTGGTATAGACACAACCGCCGTTCTTTTAAAACACCTGATGGGCAGCAGAAGCAATTACTGCCCAGATGGTATTTTTACTATCTTGGCACAGGGCCCCATGCTGGAGCCAGTTATGGAGACAGCATTGAAGGTGTCTTCTGGGTTGCAAACAGCCAAGCGGACACCAATACCCGCTCTGATATTGTCGAAAGGGACCCAAGCAGTCATGAGGCTATTCCTACTAGGTTTGCGCCCGGCACGGTATTGCCTCAGGGCTTTTATGTTGAAGGCTCTGGAAGGTCTGCACCTGCTAGCCGATCTGGTTCGCGGTCACAATCCCGTGGGCCAAATAATCGCGCTAGAAGCAGTTCCAACCAGCGCCAGCCTGCCTCTACTGTAAAACCTGATATGGCCGAAGAAATTGCTGCTCTTGTTTTGGCTAAGCTCGGTAAAGATGCCGGCCAGCCCAAGCAAGTAACGAAGCAAAGTGCCAAAGAAGTCAGGCAGAAAATTTTAAACAAGCCTCGCCAAAAGAGGACTCCAAACAAGCAGTGCCCAGTGCAGCAGTGTTTTGGAAAGAGAGGCCCCAATCAGAATTTTGGAGGCTCTGAAATGTTAAAACTTGGAACTAGTGATCCACAGTTCCCCATTCTTGCAGAGTTGGCTCCAACAGTTGGTGCCTTCTTCTTTGGATCTAAATTAGAATTGGTCAAAAAGAATTCTGGTGGTGCTGATGAACCCACCAAAGATGTGTATGAGCTGCAATATTCAGGTGCAGTTAGATTTGATAGTACTCTACCTGGTTTTGAGACTATCATGAAAGTGTTGAATGAGAATTTGAATGCCTACCAGAAGGATGGTGGTGCAGATGTGGTGAGCCCAAAGCCCCAAAGAAAAGGGCGTAGACAGGCTCAGGAAAAGAAAGATGAAGTAGATAATGTAAGCGTTGCAAAGCCCAAAAGCTCTGTGCAGCGAAATGTAAGTAGAGAATTAACCCCAGAGGATAGAAGTCTGTTGGCTCAGATCCTTGATGATGGCGTAGTGCCAGATGGGTTAGAAGATGACTCTAATGTGTAA
Protein
MSFVPGQENAGGRSSSVNRAGNGILKKTTWADQTERGPNNQNRGRRNQPKQTATTQPNSGSVVPHYSWFSGITQFQKGKEFQFAEGQGVPIANGIPASEQKGYWYRHNRRSFKTPDGQQKQLLPRWYFYYLGTGPHAGASYGDSIEGVFWVANSQADTNTRSDIVERDPSSHEAIPTRFAPGTVLPQGFYVEGSGRSAPASRSGSRSQSRGPNNRARSSSNQRQPASTVKPDMAEEIAALVLAKLGKDAGQPKQVTKQSAKEVRQKILNKPRQKRTPNKQCPVQQCFGKRGPNQNFGGSEMLKLGTSDPQFPILAELAPTVGAFFFGSKLELVKKNSGGADEPTKDVYELQYSGAVRFDSTLPGFETIMKVLNENLNAYQKDGGADVVSPKPQRKGRRQAQEKKDEVDNVSVAKPKSSVQRNVSRELTPEDRSLLAQILDDGVVPDGLEDDSNV
Summary
Function
Packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP) and plays a fundamental role during virion assembly through its interactions with the viral genome and membrane protein M. Plays an important role in enhancing the efficiency of subgenomic viral RNA transcription as well as viral replication.
Major structural component of virions that associates with genomic RNA to form a long, flexible, helical nucleocapsid. Interaction with the M protein leads to the formation of virus particles. Binds to cellular membranes and phospholipids. Elicits cell-mediated immunity. May play roles in viral transcription and translation, and/or replication. Induces transcription of the prothrombinase (FGL2) and elevates procoagulant activity.
Major structural component of virions that associates with genomic RNA to form a long, flexible, helical nucleocapsid. Interaction with the M protein leads to the formation of virus particles. Binds to cellular membranes and phospholipids. Elicits cell-mediated immunity. May play roles in viral transcription and translation, and/or replication. Induces transcription of the prothrombinase (FGL2) and elevates procoagulant activity.
Subunit
Homooligomer. Both monomeric and oligomeric forms interact with RNA. Interacts with protein M. Interacts with NSP3; this interaction serves to tether the genome to the newly translated replicase-transcriptase complex at a very early stage of infection.
Homooligomer. Both monomeric and oligomeric forms interact with RNA. Interacts with protein M. Interacts with NSP3; this interaction serves to tether the genome to the newly translated replicase-transcriptase complex at a very early stage of infection (By similarity). Interacts (when phosphorylated) with host DDX1; this interaction is essential to produce a full set of subgenomic and genomic RNAs (PubMed:25299332).
Homooligomer. Both monomeric and oligomeric forms interact with RNA. Interacts with protein M. Interacts with NSP3; this interaction serves to tether the genome to the newly translated replicase-transcriptase complex at a very early stage of infection (By similarity). Interacts (when phosphorylated) with host DDX1; this interaction is essential to produce a full set of subgenomic and genomic RNAs (PubMed:25299332).
Similarity
Belongs to the betacoronavirus nucleocapsid protein family.
Keywords
3D-structure
ADP-ribosylation
Host Golgi apparatus
Phosphoprotein
Reference proteome
Ribonucleoprotein
RNA-binding
Transcription
Transcription regulation
Viral nucleoprotein
Virion
Feature
chain Nucleoprotein
Uniprot
Q66WM6
C0KYT5
C0KYR5
C0KYY3
C0KZ04
A0A1U8QWM0
+ More
S5YA14 P03416 A0A2D1CIK8 S5YGB6 C3VPG2 Q9J3E2 P18447 Q83358 A0A513RBX1 Q83360 H9BZY3 D7PJY5 P18448 D4QFK6 A0A2S0SZ21 Q5ICW7 Q9WCD0 C0KYZ4 C0KYS6 C0KYU5 I7B999 C0KYV6 I7B990 P03417 Q08614 C0KYX4 A7U549 C0KYW6 P18446 Q77NL5 Q9PY96 Q02915 Q9QCZ9 Q83359 Q9J3F6 I1TMG3 P59713 C6GHS6 Q83357 V5N735 I1TMH3 A0A096XNI2 A0A2H4N014 A0A2H4MXD4 A0A096XNH5 A0A096XNH4 A0A2H4MX28 A0A3G3NHM6 A0A3G3NH02 A8R4D8 Q9DQX6 I2FKH0 A0A4P2V418 E5RPZ1 A0A2H4MWZ7 R4WKE3 A0A4P2V3N3 A0A0K2RVN9 A0A0M5NBL9 A0A142D7Z2 A0A166IK31 A0A1W6DZX0 A0A5J6NU34 Q2QKM8 A0A5J6NRV2 B7TYH5 A0A3T0IBL9 Q8JSP4 Q9QAR1 B7U2S1 C6GHR6 C6GHL9 H9AA69 Q0PL33 A0A142D800 A0A142D7Z3 B7U2R0 P26020 A0A2H4MZ40 A0A2R4STW5 A0A142D7Z0 A0A142D801 A0A166MX98 Q6W358 A3E2F7 B8YPE5 A0A5J6NS08 Q9QAR8 A0A410UD36 A0A410UD67 H9AA48 B8RIR3 A7BKC8
S5YA14 P03416 A0A2D1CIK8 S5YGB6 C3VPG2 Q9J3E2 P18447 Q83358 A0A513RBX1 Q83360 H9BZY3 D7PJY5 P18448 D4QFK6 A0A2S0SZ21 Q5ICW7 Q9WCD0 C0KYZ4 C0KYS6 C0KYU5 I7B999 C0KYV6 I7B990 P03417 Q08614 C0KYX4 A7U549 C0KYW6 P18446 Q77NL5 Q9PY96 Q02915 Q9QCZ9 Q83359 Q9J3F6 I1TMG3 P59713 C6GHS6 Q83357 V5N735 I1TMH3 A0A096XNI2 A0A2H4N014 A0A2H4MXD4 A0A096XNH5 A0A096XNH4 A0A2H4MX28 A0A3G3NHM6 A0A3G3NH02 A8R4D8 Q9DQX6 I2FKH0 A0A4P2V418 E5RPZ1 A0A2H4MWZ7 R4WKE3 A0A4P2V3N3 A0A0K2RVN9 A0A0M5NBL9 A0A142D7Z2 A0A166IK31 A0A1W6DZX0 A0A5J6NU34 Q2QKM8 A0A5J6NRV2 B7TYH5 A0A3T0IBL9 Q8JSP4 Q9QAR1 B7U2S1 C6GHR6 C6GHL9 H9AA69 Q0PL33 A0A142D800 A0A142D7Z3 B7U2R0 P26020 A0A2H4MZ40 A0A2R4STW5 A0A142D7Z0 A0A142D801 A0A166MX98 Q6W358 A3E2F7 B8YPE5 A0A5J6NS08 Q9QAR8 A0A410UD36 A0A410UD67 H9AA48 B8RIR3 A7BKC8
Pubmed
8392595
10438865
15709029
17079303
16341254
6687635
+ More
2171216 9426441 8395114 12594208 17367888 17210170 23760243 28591694 28720894 29199039 19782089 11582515 7733827 20519394 15507445 10551451 12970410 12663803 6308569 25299332 8390823 20631137 17041219 11502093 10187767 8438589 10882653 1312608 25463600 30285857 17706262 11101590 21273011 26271151 23648375 26395082 26969241 27084706 31534035 16809333 12237422 9778786 18842722 22398294 1856695 17092595 19162098 17374765
2171216 9426441 8395114 12594208 17367888 17210170 23760243 28591694 28720894 29199039 19782089 11582515 7733827 20519394 15507445 10551451 12970410 12663803 6308569 25299332 8390823 20631137 17041219 11502093 10187767 8438589 10882653 1312608 25463600 30285857 17706262 11101590 21273011 26271151 23648375 26395082 26969241 27084706 31534035 16809333 12237422 9778786 18842722 22398294 1856695 17092595 19162098 17374765
EMBL
FJ884686
ACO72888.1
FJ647220
ACN89710.1
FJ647218
ACN89686.1
+ More
FJ647225 ACN89754.1 FJ647227 ACN89774.1 AY700211 MF618252 AAU06361.1 ATN37892.1 KF268336 KF268338 KF268339 AGT17721.1 AGT17743.1 AGT17754.1 X00509 M35256 AF029248 S64884 MF618253 ATN37902.1 KF268337 AGT17732.1 FJ884687 ACO72897.1 AF208067 AAF69349.1 M35254 L37758 AAA74733.1 MH687883 QCI55924.1 L37760 JQ173883 AFD97608.1 GU593319 ADI59793.1 M35255 AB551247 BAJ04701.1 MF416379 AWB14620.1 AY771998 AAW47245.1 AF088984 FJ647226 ACN89761.1 FJ647219 ACN89697.1 FJ647221 ACN89716.1 JX169867 AFO11522.1 FJ647222 ACN89733.1 JX169866 AFO11512.1 X00990 X63538 CAA45099.1 FJ647224 ACN89747.1 EF682498 ABS87269.1 FJ647223 ACN89735.1 M35253 AF208066 AAF69338.1 AF201929 AF061835 D10760 AF207551 AF190408 AAF05706.1 L37759 AAA76578.1 AF207902 AAF68926.1 JF792616 AFG25751.1 AJ544718 FJ938068 ACT11046.1 M80644 AAA46462.1 KF850449 AHA83612.1 JF792617 AFG25761.1 KF294372 AID16664.1 KY370049 ATP66765.1 KY370048 ATP66759.1 KF294371 AID16658.1 KF294370 AID16652.1 KY370051 ATP66776.1 MH687968 AYR18603.1 MH687970 AYR18621.1 EF446615 ABP87995.1 AF251144 AB671298 BAM15655.1 LC149486 BBD43533.1 AB555559 LC061272 BAJ52884.1 BAS18850.1 KY370046 ATP66747.1 AB775893 LC061274 LC054263 BAN18702.1 BAS18870.1 BAS53429.1 LC149485 BBD43532.1 LC061273 BAS18860.1 LC054264 BAS53430.1 KT318085 AMQ23461.1 KT852998 ANA11064.1 KY967359 ARK08665.1 MN514963 QEY10637.1 DQ011855 AAY68302.1 MN514966 QEY10661.1 FJ415324 ACJ35489.1 MH810163 AZU96330.1 AF481863 AF058943 FJ425189 ACJ67006.1 FJ938067 ACT11036.1 FJ938063 ACT10989.1 JN874562 AFE48831.1 DQ811784 ABG89284.1 KT318093 AMQ23469.1 KT318086 KT318088 AMQ23462.1 AMQ23464.1 FJ425188 FJ425190 ACJ66985.1 AF058944 KY370052 ATP66782.1 MH043952 AVZ61105.1 KT318083 AMQ23459.1 KT318089 KT318090 KT318091 KT318094 KT318095 KX982264 MG757140 MG757141 MG757142 AMQ23465.1 AMQ23466.1 AMQ23467.1 AMQ23470.1 AMQ23471.1 ARD06005.1 AVI15028.1 AVI15039.1 AVI15050.1 KU127229 KY994645 MF083115 ANA52021.1 AUF40280.1 AVV64339.1 AY316299 AAQ67202.1 DQ682406 ABG78753.1 FJ556872 ACL80212.1 MN514964 MN514965 MN514967 QEY10645.1 QEY10653.1 QEY10669.1 AF058942 MH475352 QAU20980.1 MH475353 QAU20981.1 JN874560 AFE48809.1 EU999954 ACL13001.1 AB354579 BAF75637.1
FJ647225 ACN89754.1 FJ647227 ACN89774.1 AY700211 MF618252 AAU06361.1 ATN37892.1 KF268336 KF268338 KF268339 AGT17721.1 AGT17743.1 AGT17754.1 X00509 M35256 AF029248 S64884 MF618253 ATN37902.1 KF268337 AGT17732.1 FJ884687 ACO72897.1 AF208067 AAF69349.1 M35254 L37758 AAA74733.1 MH687883 QCI55924.1 L37760 JQ173883 AFD97608.1 GU593319 ADI59793.1 M35255 AB551247 BAJ04701.1 MF416379 AWB14620.1 AY771998 AAW47245.1 AF088984 FJ647226 ACN89761.1 FJ647219 ACN89697.1 FJ647221 ACN89716.1 JX169867 AFO11522.1 FJ647222 ACN89733.1 JX169866 AFO11512.1 X00990 X63538 CAA45099.1 FJ647224 ACN89747.1 EF682498 ABS87269.1 FJ647223 ACN89735.1 M35253 AF208066 AAF69338.1 AF201929 AF061835 D10760 AF207551 AF190408 AAF05706.1 L37759 AAA76578.1 AF207902 AAF68926.1 JF792616 AFG25751.1 AJ544718 FJ938068 ACT11046.1 M80644 AAA46462.1 KF850449 AHA83612.1 JF792617 AFG25761.1 KF294372 AID16664.1 KY370049 ATP66765.1 KY370048 ATP66759.1 KF294371 AID16658.1 KF294370 AID16652.1 KY370051 ATP66776.1 MH687968 AYR18603.1 MH687970 AYR18621.1 EF446615 ABP87995.1 AF251144 AB671298 BAM15655.1 LC149486 BBD43533.1 AB555559 LC061272 BAJ52884.1 BAS18850.1 KY370046 ATP66747.1 AB775893 LC061274 LC054263 BAN18702.1 BAS18870.1 BAS53429.1 LC149485 BBD43532.1 LC061273 BAS18860.1 LC054264 BAS53430.1 KT318085 AMQ23461.1 KT852998 ANA11064.1 KY967359 ARK08665.1 MN514963 QEY10637.1 DQ011855 AAY68302.1 MN514966 QEY10661.1 FJ415324 ACJ35489.1 MH810163 AZU96330.1 AF481863 AF058943 FJ425189 ACJ67006.1 FJ938067 ACT11036.1 FJ938063 ACT10989.1 JN874562 AFE48831.1 DQ811784 ABG89284.1 KT318093 AMQ23469.1 KT318086 KT318088 AMQ23462.1 AMQ23464.1 FJ425188 FJ425190 ACJ66985.1 AF058944 KY370052 ATP66782.1 MH043952 AVZ61105.1 KT318083 AMQ23459.1 KT318089 KT318090 KT318091 KT318094 KT318095 KX982264 MG757140 MG757141 MG757142 AMQ23465.1 AMQ23466.1 AMQ23467.1 AMQ23470.1 AMQ23471.1 ARD06005.1 AVI15028.1 AVI15039.1 AVI15050.1 KU127229 KY994645 MF083115 ANA52021.1 AUF40280.1 AVV64339.1 AY316299 AAQ67202.1 DQ682406 ABG78753.1 FJ556872 ACL80212.1 MN514964 MN514965 MN514967 QEY10645.1 QEY10653.1 QEY10669.1 AF058942 MH475352 QAU20980.1 MH475353 QAU20981.1 JN874560 AFE48809.1 EU999954 ACL13001.1 AB354579 BAF75637.1
Proteomes
UP000180349
UP000098534
UP000151686
UP000119015
UP000138757
UP000113498
+ More
UP000112338 UP000164259 UP000171975 UP000007192 UP000171340 UP000180350 UP000155907 UP000118088 UP000117254 UP000154289 UP000148943 UP000158592 UP000133456 UP000149957 UP000142874 UP000114687 UP000007193 UP000167944 UP000159025 UP000143567 UP000139707 UP000163097 UP000175084 UP000173347 UP000161770 UP000103372 UP000166748 UP000155347 UP000130220 UP000125924 UP000158296 UP000007543 UP000133443 UP000164707 UP000172225 UP000145001 UP000147981 UP000102696 UP000148210 UP000124585 UP000168103
UP000112338 UP000164259 UP000171975 UP000007192 UP000171340 UP000180350 UP000155907 UP000118088 UP000117254 UP000154289 UP000148943 UP000158592 UP000133456 UP000149957 UP000142874 UP000114687 UP000007193 UP000167944 UP000159025 UP000143567 UP000139707 UP000163097 UP000175084 UP000173347 UP000161770 UP000103372 UP000166748 UP000155347 UP000130220 UP000125924 UP000158296 UP000007543 UP000133443 UP000164707 UP000172225 UP000145001 UP000147981 UP000102696 UP000148210 UP000124585 UP000168103
Pfam
PF00937
Corona_nucleoca
ProteinModelPortal
Q66WM6
C0KYT5
C0KYR5
C0KYY3
C0KZ04
A0A1U8QWM0
+ More
S5YA14 P03416 A0A2D1CIK8 S5YGB6 C3VPG2 Q9J3E2 P18447 Q83358 A0A513RBX1 Q83360 H9BZY3 D7PJY5 P18448 D4QFK6 A0A2S0SZ21 Q5ICW7 Q9WCD0 C0KYZ4 C0KYS6 C0KYU5 I7B999 C0KYV6 I7B990 P03417 Q08614 C0KYX4 A7U549 C0KYW6 P18446 Q77NL5 Q9PY96 Q02915 Q9QCZ9 Q83359 Q9J3F6 I1TMG3 P59713 C6GHS6 Q83357 V5N735 I1TMH3 A0A096XNI2 A0A2H4N014 A0A2H4MXD4 A0A096XNH5 A0A096XNH4 A0A2H4MX28 A0A3G3NHM6 A0A3G3NH02 A8R4D8 Q9DQX6 I2FKH0 A0A4P2V418 E5RPZ1 A0A2H4MWZ7 R4WKE3 A0A4P2V3N3 A0A0K2RVN9 A0A0M5NBL9 A0A142D7Z2 A0A166IK31 A0A1W6DZX0 A0A5J6NU34 Q2QKM8 A0A5J6NRV2 B7TYH5 A0A3T0IBL9 Q8JSP4 Q9QAR1 B7U2S1 C6GHR6 C6GHL9 H9AA69 Q0PL33 A0A142D800 A0A142D7Z3 B7U2R0 P26020 A0A2H4MZ40 A0A2R4STW5 A0A142D7Z0 A0A142D801 A0A166MX98 Q6W358 A3E2F7 B8YPE5 A0A5J6NS08 Q9QAR8 A0A410UD36 A0A410UD67 H9AA48 B8RIR3 A7BKC8
S5YA14 P03416 A0A2D1CIK8 S5YGB6 C3VPG2 Q9J3E2 P18447 Q83358 A0A513RBX1 Q83360 H9BZY3 D7PJY5 P18448 D4QFK6 A0A2S0SZ21 Q5ICW7 Q9WCD0 C0KYZ4 C0KYS6 C0KYU5 I7B999 C0KYV6 I7B990 P03417 Q08614 C0KYX4 A7U549 C0KYW6 P18446 Q77NL5 Q9PY96 Q02915 Q9QCZ9 Q83359 Q9J3F6 I1TMG3 P59713 C6GHS6 Q83357 V5N735 I1TMH3 A0A096XNI2 A0A2H4N014 A0A2H4MXD4 A0A096XNH5 A0A096XNH4 A0A2H4MX28 A0A3G3NHM6 A0A3G3NH02 A8R4D8 Q9DQX6 I2FKH0 A0A4P2V418 E5RPZ1 A0A2H4MWZ7 R4WKE3 A0A4P2V3N3 A0A0K2RVN9 A0A0M5NBL9 A0A142D7Z2 A0A166IK31 A0A1W6DZX0 A0A5J6NU34 Q2QKM8 A0A5J6NRV2 B7TYH5 A0A3T0IBL9 Q8JSP4 Q9QAR1 B7U2S1 C6GHR6 C6GHL9 H9AA69 Q0PL33 A0A142D800 A0A142D7Z3 B7U2R0 P26020 A0A2H4MZ40 A0A2R4STW5 A0A142D7Z0 A0A142D801 A0A166MX98 Q6W358 A3E2F7 B8YPE5 A0A5J6NS08 Q9QAR8 A0A410UD36 A0A410UD67 H9AA48 B8RIR3 A7BKC8
PDB
3HD4
E-value=1.63683e-72,
Score=694
Ontologies
KEGG
GO
Subcellular Location
From MSLVP
Host nucleus > host nucleolus. Host cytoplasm. Secreted.
From Uniprot
Host Golgi apparatus
Located inside the virion, complexed with the viral RNA. Probably associates with ER-derived membranes where it participates in viral RNA synthesis and virus budding. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment Located inside the virion, complexed with the viral RNA. Probably associates with ER-derived membranes where it participates in viral RNA synthesis and virus budding. With evidence from 1 publications.
Virion Located inside the virion, complexed with the viral RNA. Probably associates with ER-derived membranes where it participates in viral RNA synthesis and virus budding. With evidence from 1 publications.
Host endoplasmic reticulum-Golgi intermediate compartment Located inside the virion, complexed with the viral RNA. Probably associates with ER-derived membranes where it participates in viral RNA synthesis and virus budding. With evidence from 1 publications.
Virion Located inside the virion, complexed with the viral RNA. Probably associates with ER-derived membranes where it participates in viral RNA synthesis and virus budding. With evidence from 1 publications.
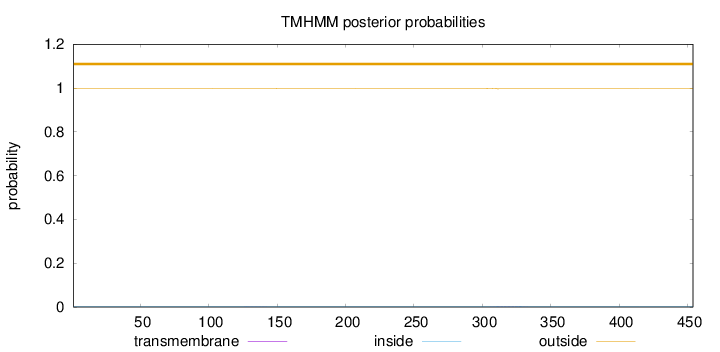
Topology
Length:
454
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04682
Exp number, first 60 AAs:
6e-05
Total prob of N-in:
0.00165
outside
1 - 454
Population Genetic Test Statistics
Genomic alignment in the CDS region
Multiple alignment of Orthologues
Gene Tree
Orthologous in Strains
| Strain | Availability | Status | Gene |
|---|---|---|---|
| CHINA_HS_2019_MN908947 | |||
| CHINA_AVIAN_2008_NC_016995 | |||
| CHINA_AVIAN_2007_NC_016991 | |||
| CHINA_MURINE_2015_NC_035191 | |||
| CANADA_AVIAN_2007_NC_010800 | |||
| USA_PIG_2000_NC_038861 | |||
| CHINA_AVIAN_2007_NC_011549 | |||
| ITALY_PIG_2009_NC_028806 | |||
| GERMANY_PIG_2012_LT545990 | |||
| CHINA_AVIAN_2007_NC_016992 | |||
| CHINA_BAT_2005_NC_009657 | |||
| CANADA_HS_2003_NC_004718 | |||
| CHINA_BAT_2014_NC_030886 | |||
| CHINA_BAT_2005_NC_018871 | |||
| USA_MURINE_2009_NC_012936 |
Protein |
YP_003029852.1 |
|
| CHINA_RABBIT_2006_NC_017083 | |||
| UK_PIG_2000_NC_003436 | |||
| ROMANIA_PIG_2015_LT898435 | |||
| ROMANIA_PIG_2015_LT898436 | |||
| GERMANY_PIG_2015_LT898444 | |||
| GERMANY_PIG_2015_LT898414 | |||
| GERMANY_PIG_2015_LT898439 | |||
| GERMANY_PIG_2015_LT898413 | |||
| GERMANY_PIG_2015_LT898412 | |||
| GERMANY_PIG_2015_LT898411 | |||
| GERMANY_PIG_2015_LT898416 | |||
| GERMANY_PIG_2015_LT898443 | |||
| GERMANY_PIG_2015_LT898420 | |||
| GERMANY_PIG_2015_LT898408 | |||
| GERMANY_PIG_2015_LT898423 | |||
| GERMANY_PIG_2015_LT898432 | |||
| GERMANY_PIG_2015_LT898409 | |||
| GERMANY_PIG_2015_LT898425 | |||
| GERMANY_PIG_2015_LT898446 | |||
| GERMANY_PIG_2014_LT898438 | |||
| GERMANY_PIG_2014_LT898440 | |||
| GERMANY_PIG_2014_LT900501 | |||
| GERMANY_PIG_2014_LT898427 | |||
| GERMANY_PIG_2014_LT898415 | |||
| GERMANY_PIG_2014_LT898421 | |||
| GERMANY_PIG_2014_LT898431 | |||
| GERMANY_PIG_2014_LT898410 | |||
| GERMANY_PIG_2014_LT900498 | |||
| GERMANY_PIG_2014_LT898430 | |||
| GERMANY_PIG_1978_LT897799 | |||
| GERMANY_PIG_2014_LT898447 | |||
| GERMANY_PIG_2014_LT898426 | |||
| GERMANY_PIG_2014_LT898417 | |||
| GERMANY_PIG_2014_LT900500 | |||
| GERMANY_PIG_2014_LT898445 | |||
| AUSTRIA_PIG_2015_LT898418 | |||
| AUSTRIA_PIG_2015_LT898441 | |||
| AUSTRIA_PIG_2015_LT898433 | |||
| AUSTRIA_PIG_2015_LT900502 | |||
| GERMANY_PIG_2015_LT900499 | |||
| BELGIUM_PIG_1980_LT906620 | |||
| BELGIUM_PIG_1977_LT905450 | |||
| SWITZERLAND_PIG_2003_LT905451 | |||
| BELGIUM_PIG_1978_LT906581 | |||
| UK_PIG_1987_LT906582 | |||
| CHINA_PIG_2009_NC_016990 | |||
| CHINA_PIG_2010_NC_039208 | |||
| KENYA_BAT_2010_KY073745 | |||
| KENYA_BAT_2010_NC_032107 | |||
| CHINA_AVIAN_2007_NC_016994 | |||
| EUROPE_MURINE_2004_AY700211 |
Protein |
AAU06361.1 |
|
| CHINA_AVIAN_2007_NC_011550 | |||
| USA_MURINE_1997_NC_001846 |
Protein |
NP_045302.1 |
|
| USA_MURINE_1998_NC_023760 | |||
| MID_EAST_HS_2012_NC_019843 | |||
| CHINA_AVIAN_2007_NC_016993 | |||
| CHINA_MURINE_2013_NC_032730 | |||
| USA_HS_2004_AY585228 |
Protein |
AAT84358.1 |
|
| NETHERLAND_HS_2004_NC_005831 | |||
| CHINA_HS_2004_NC_006577 |
Protein |
YP_173242.1 |
|
| EUROPE_HS_2000_NC_002645 | |||
| NETHERLAND_FERRET_2010_NC_030292 | |||
| JAPAN_FERRET_2013_LC119077 | |||
| USA_FELINE_2005_NC_002306 | |||
| CHINA_MURINE_2011_NC_034972 | |||
| CHINA_AVIAN_2007_NC_016996 | |||
| ARABIA_CAMEL_2015_NC_028752 | |||
| CHINA_AVIAN_2007_NC_011547 | |||
| CHINA_BAT_2012_NC_028824 | |||
| CHINA_BAT_2013_NC_028814 | |||
| CHINA_BAT_2013_NC_028833 | |||
| CHINA_BAT_2011_NC_028811 | |||
| USA_BOVIN_2001_NC_003045 |
Protein |
NP_150083.1 |
|
| CHINA_MURINE_2012_NC_026011 |
Protein |
YP_009113031.1 |
|
| GERMANY_ERINACEINAE_2012_NC_022643 | |||
| GERMANY_ERINACEINAE_2012_NC_039207 | |||
| UK_HS_2012_NC_038294 | |||
| AMERICA_WHALE_2007_NC_010646 | |||
| CHINA_BAT_2013_NC_025217 | |||
| UGANDA_BAT_2013_NC_034440 | |||
| CHINA_BAT_2006_NC_009021 | |||
| CHINA_BAT_2008_NC_010438 | |||
| CHINA_BAT_2006_NC_009020 | |||
| CHINA_BAT_2006_NC_009019 | |||
| CHINA_BAT_2006_NC_009988 | |||
| USA_BAT_2006_NC_022103 | |||
| BULGARIA_BAT_2008_NC_014470 | |||
| CHINA_BAT_2008_NC_010437 | |||
| USA_AVIAN_2004_NC_001451 |
Copyright@ 2018-2023
Any Comments and suggestions mail to:
zhuzl@cqu.edu.cn,
mg@cau.edu.cn
渝ICP备19006517号


渝公网安备 50010602502065号
In processing...
Login to ASFVdb
Email
Password
Please go to Regist if without an account.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
If you have forgotten your password, you can once again Regist an account with a registed or new email.
Change my password
Enter new password
Reenter new password
Regist an account of ASFVdb
It is required that you provide your institutional e-mail address (with edu or org in the domain) as confirmation of your affiliation.
Enter email
Reenter email
First Name
Last Name
Institution
You can directly go to Login if with an account.
Registraion Success
Your password has been sent to your email.
Please check it and login later.
Welcome to use ASFVdb.
Please check it and login later.
Welcome to use ASFVdb.